DISCUSSION
HCC is currently the most common type of primary liver cancer in adults. It is also the most frequent cause of death in patients with liver cirrhosis. HCC in many countries, including Korea, is closely related to HBV infection. Although patients infected with HBV are regularly screened for HCC by measuring blood levels of alpha-fetoprotein (AFP) and abdominal ultrasonography every six months, HBV-associated HCC remains the main cause of mortality in HBV-infected patients [
4]. The identification of new biomarkers and regulatory pathways related to the development of HCC in HBV-infected patients can enhance early detection, diagnosis, and treatment of HCC. The present study utilized bioinformatics to analyze the GSE121248 gene expression dataset and screen for key genes and molecular pathways that are closely related to the development of HCC. Using cut-off criteria of
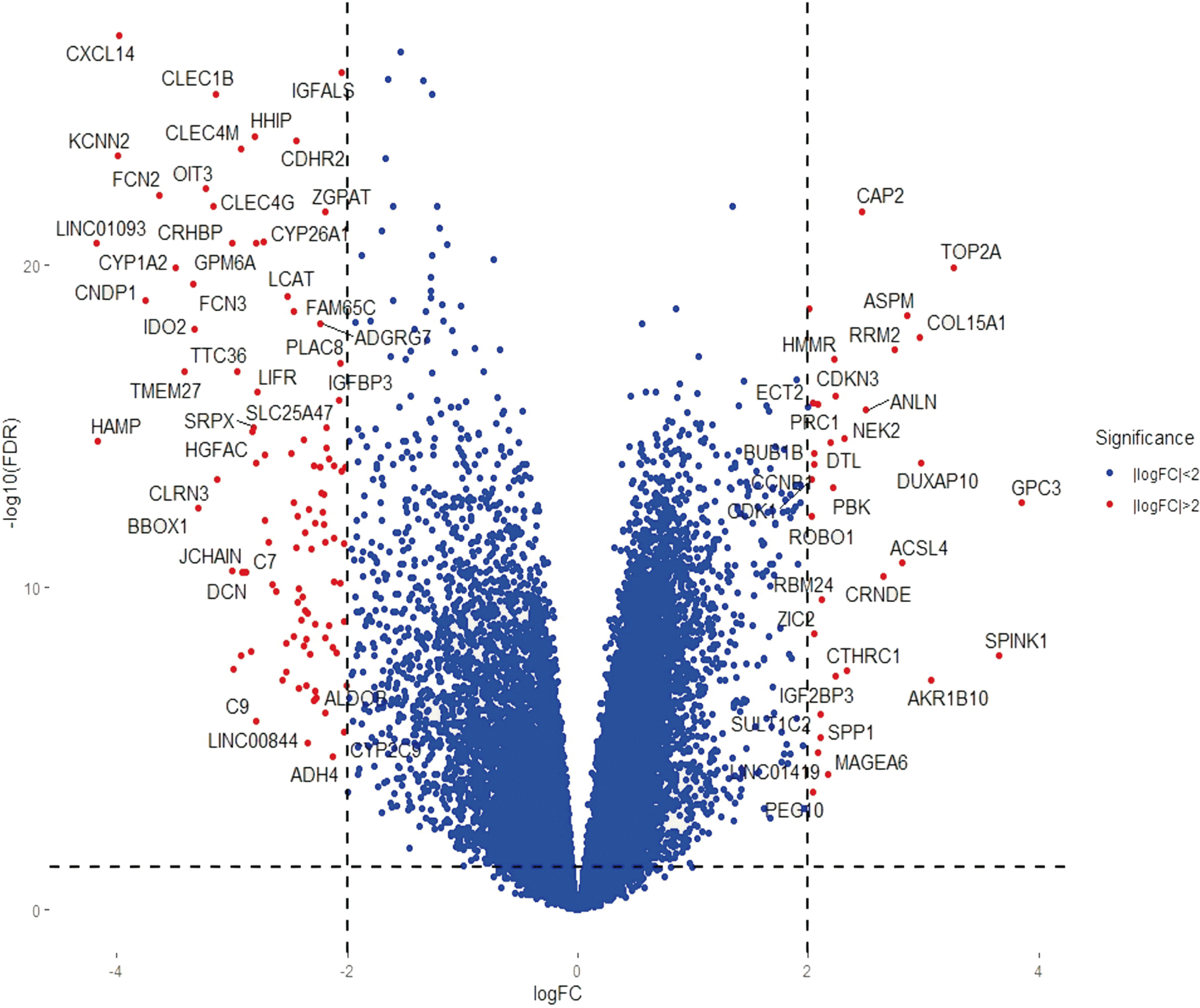
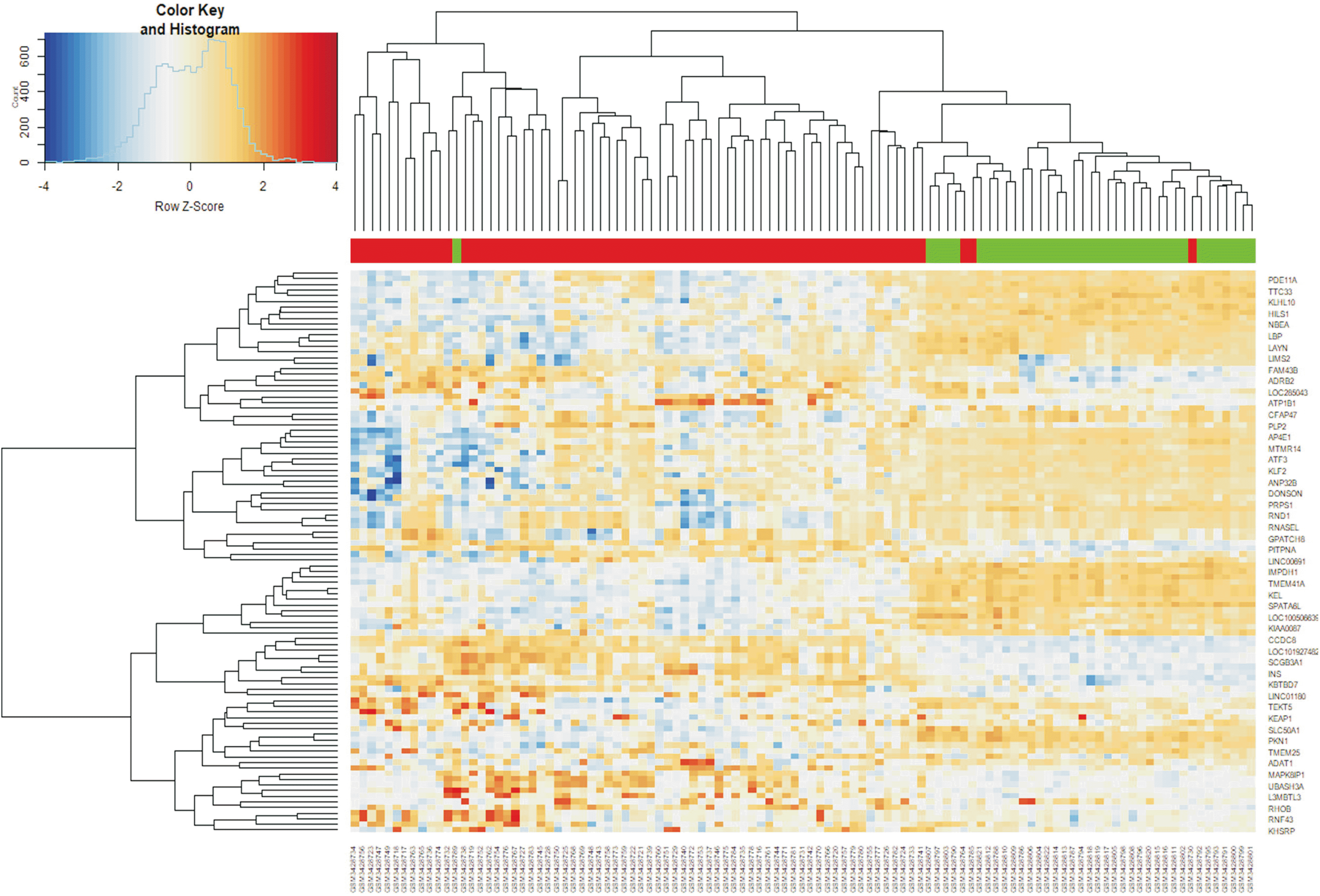
p < 0.05 and | log
2 FC | > 2, this analysis identified 134 DEGs, of which 100 were down-regulated and 34 were up-regulated in HCC than in normal liver tissues. To obtain a comprehensive understanding of these DEGs, we performed bioinformatics analyses, including GO enrichment, KEGG pathway, and PPI network analyses. To validate the 14 hub genes identified, we also analyzed their expression levels using the GEPIA website and assessed their effects on overall patient survival using the TCGA database.
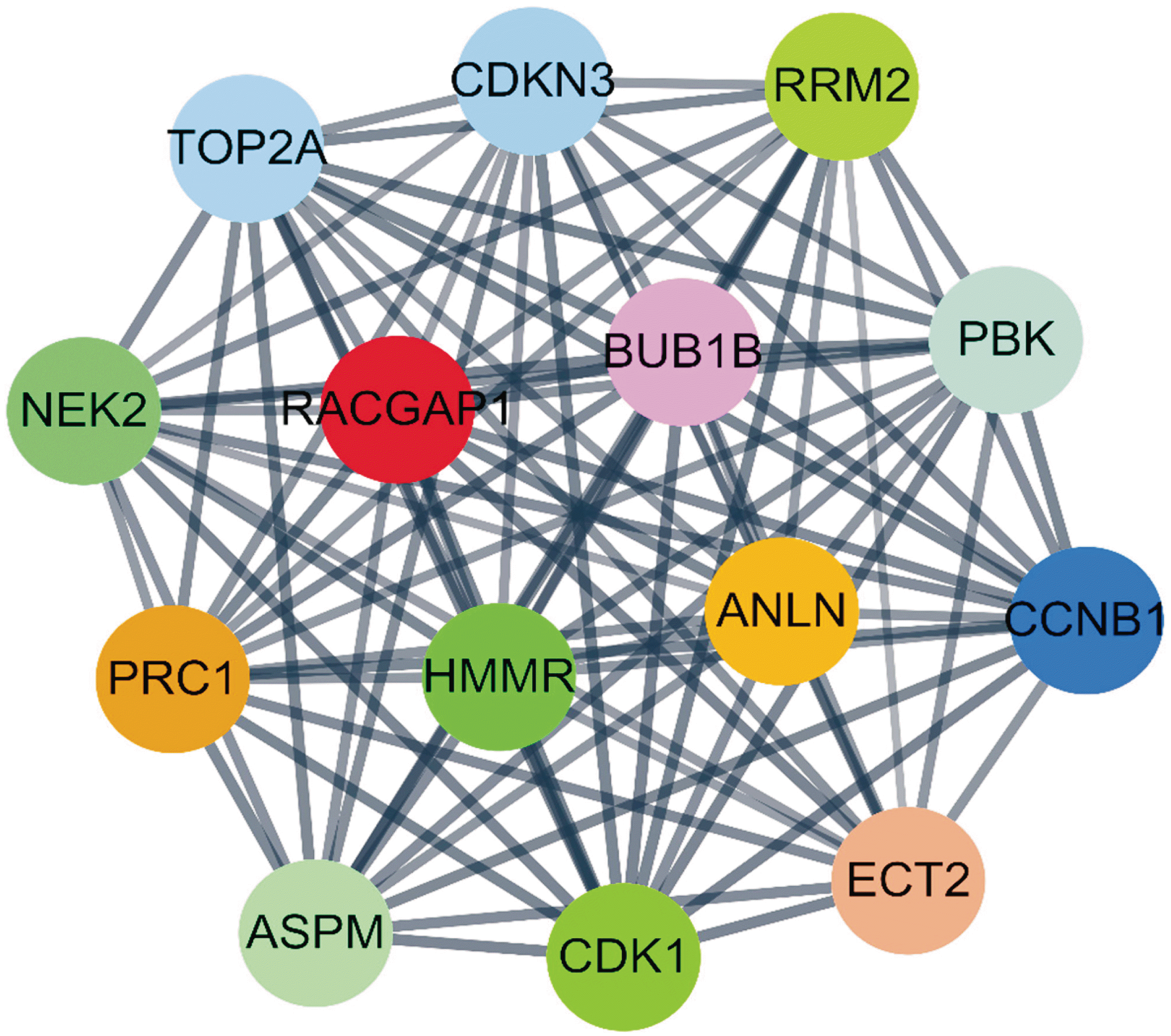
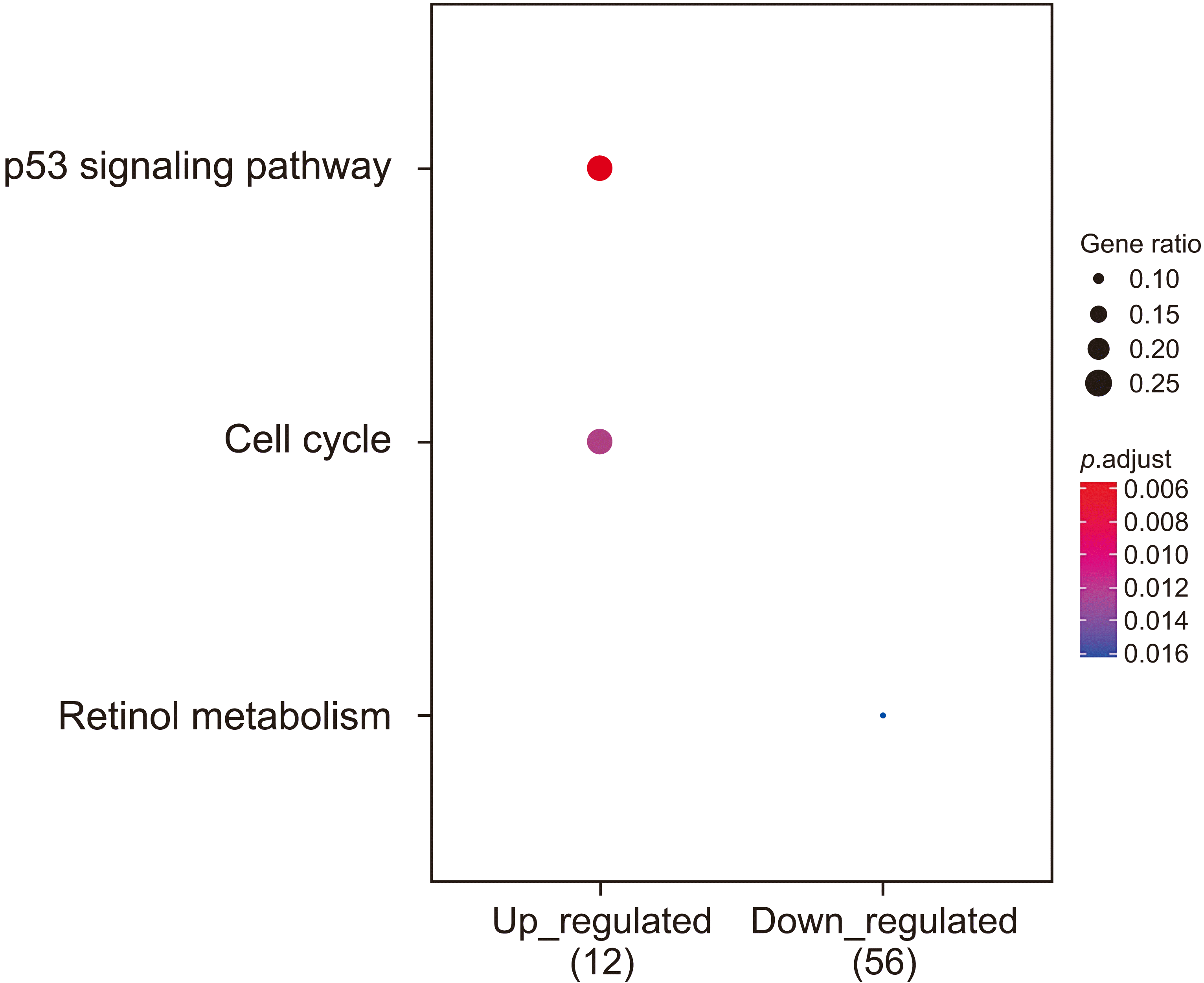
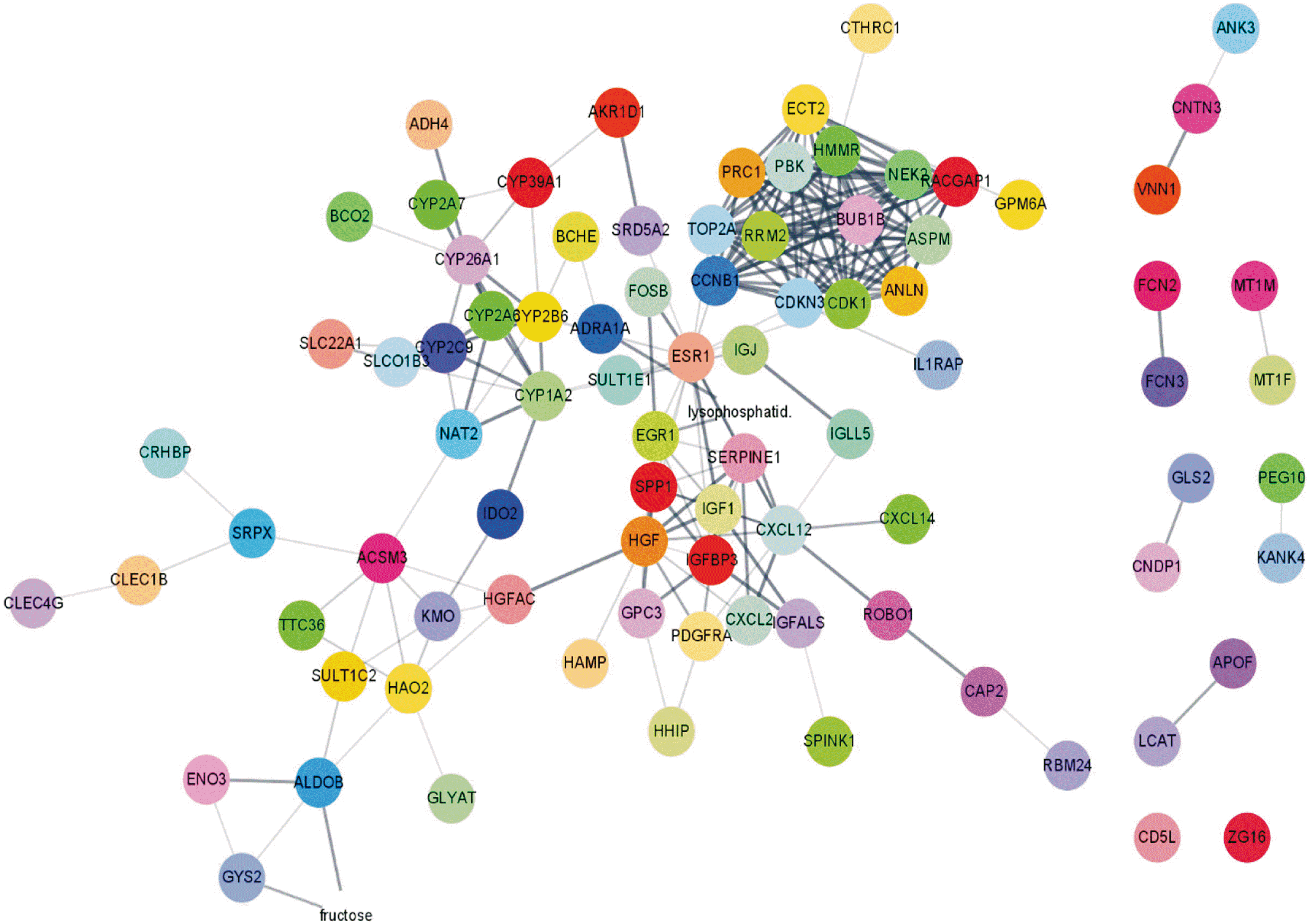
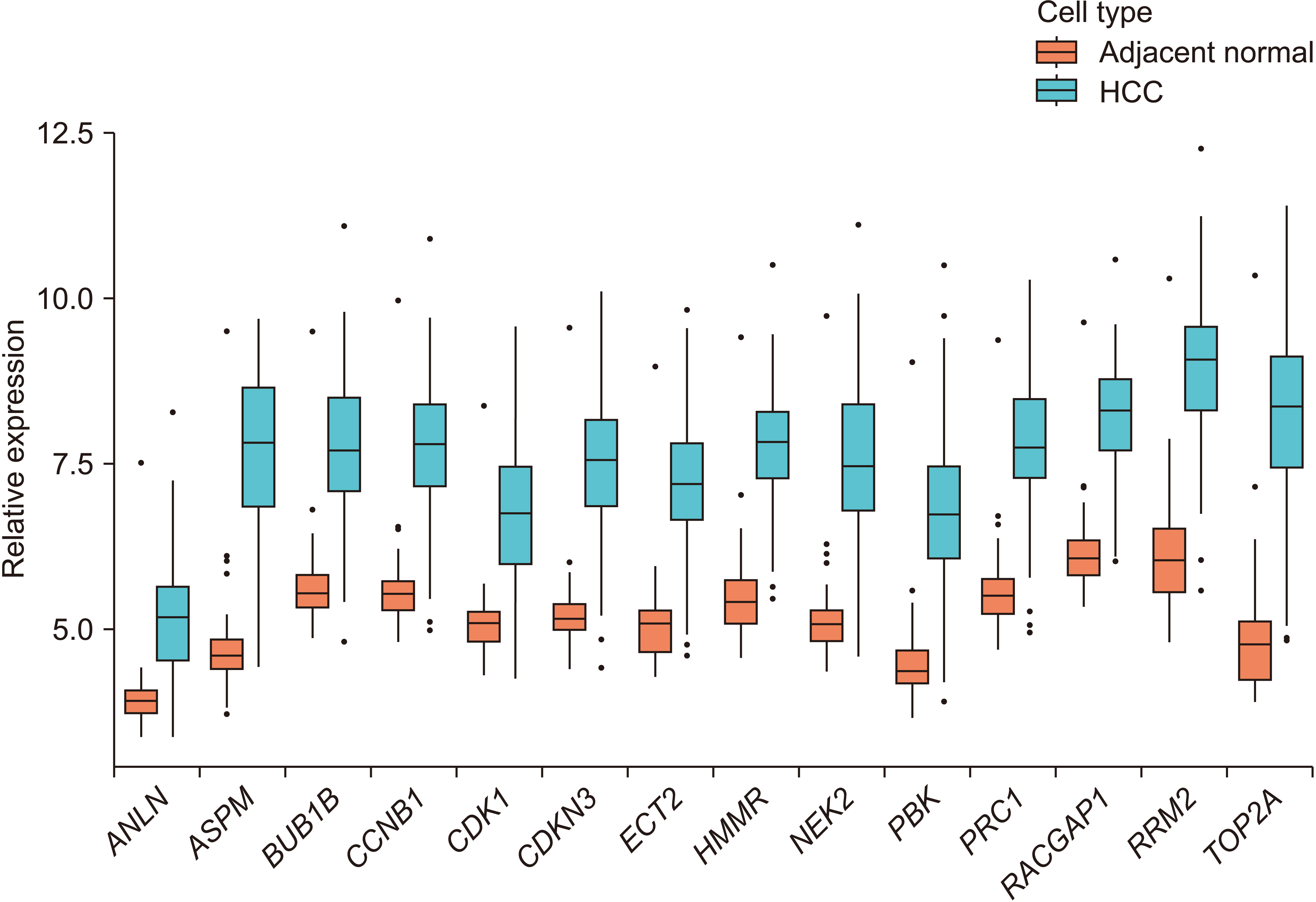
GO enrichment analysis revealed that the 34 up-regulated DEGs were significantly enriched in genes involved in nuclear division and organelle fission, whereas the 100 down-regulated DEGs were highly enriched in genes involved in steroid metabolism. KEGG pathway analysis showed that the 34 up-regulated genes were enriched in genes involved in the p53 signaling pathway, whereas the 100 down-regulated genes were significantly enriched in genes involved in retinol metabolism. PPI network analysis was performed using STRING. Hub genes were identified by MCODE, a Cytoscape plug-in. The 14 hub genes identified by PPI network analysis included ANLN, ASPM, BUB1B, CCNB1, CDK1, CDKN3, ECT2, HMMR, NEK2, PBK, PRC1, RACGAP1, RRM2, and TOP2A. Analysis using the GEPIA website showed that expression levels of all 14 mRNAs were much higher in HCC than in normal tissue samples. Survival analysis using the TCGA database showed that expression levels of these genes were significantly associated with patient survival.
Anillin (ANLN) is a conserved protein implicated in cytoskeletal dynamics during cytokinesis and cellularization. ANLN is essential for cell division. Therefore, it is critical for development and homeostasis in metazoans. ANLN expression levels are correlated with the metastatic potential of human tumors. For example, colorectal cancer cell lines expressing higher levels of ANLN replicated faster, were more invasive, and had greater levels of migration than cell lines expressing lower levels of ANLN [
5]. Overexpression of ANLN may enhance cell migration by cytoskeletal remodeling, promoting the proliferation, invasion, and motility of tumor cells [
6]. In addition, overexpression of ANLN involving copy number variation, as well as alterations in DNA and histone methylation, was found to be related to poor prognosis in patients with HCC [
7].
The abnormal spindle-like microcephaly-associated (ASPM) protein plays a significant role in neurogenesis and cell proliferation. In addition, ASPM overexpression has been associated with poor outcomes of patients with various cancers [
8,
9]. ASPM overexpression may predict HCC invasiveness or metastatic potential. It is associated with a higher risk of early tumor recurrence regardless of p53 mutation status or tumor stage [
10].
The mitotic checkpoint serine/threonine-protein kinase BUB1 beta (
BUB1B) gene encodes a kinase involved in spindle checkpoint function and chromosome segregation. Impaired spindle checkpoint function is involved in the initiation and development of tumors. In addition, BUB1B can promote HCC progression by activating the mTORC1 signaling pathway [
11].
Cyclin B1 (CCNB1) is a regulatory protein involved in mitosis. A high level of CCNB1 expression may be associated with the extent of tumor progression and invasion, suggesting that the expression level of CCNB1 might be prognostic of survival outcomes in cancer patients. Transcriptional activation of CCNB1 is essential for the proliferation of HCC. It is closely associated with poor prognosis in patients with HCC [
12].
Cyclin-dependent kinase 1 (CDK1), a member of the Ser/Thr protein kinase family, plays an important role in G1/S and G2/M phase transitions of eukaryotic cell cycle by interacting with CCNB1. CDK1 was found to play an essential role in HCC development by regulating subcellular localization of apoptin [
13]. Metformin can significantly inhibit HCC cell proliferation by inducing G2/M arrest and reducing the expression of CDK1, suggesting that CDK1 might be involved in the process of HCC cell proliferation [
14]. In addition, overexpression of CDK1 in HCC is significantly associated with large tumor size, advanced tumor stage, portal invasion, intrahepatic metastasis, poor differentiation, high AFP level, high Ki-67 index, and poor patient prognosis [
15].
Cyclin-dependent kinase inhibitor 3 (CDKN3), which inhibits the activation of cyclin-dependent kinase 2 (CDK2), can promote tumorigenesis in various types of cancer [
16]. In contrast to our findings, a recent study has found that CDKN3 is negatively associated with pathological tumor stage and that its inhibition can enhance HCC cell survival [
17].
The epithelial cell transformation sequence 2 (ECT2) protein is a Rho-specific exchange factor. Activation of the ECT2/Rho pathway can enhance the progression of several tumors. ECT2 overexpression may promote the progression and early recurrence of HCC by regulating the Rho/ERK signaling axis [
18]. High levels of ECT2 have been implicated in other types of cancer [
19].
Hyaluronan-mediated motility receptor (HMMR or RHAMM) was originally identified as a soluble protein that altered migratory cell behavior and bound to hyaluronan. HMMR expression is associated with breast cancer risk and the progression of several tumor types. A recent bioinformatics study has found that overexpression of HMMR is positively correlated with HCC tumor grade, pathological stage, and tumor stage [
20].
The serine/threonine-protein kinase Nek2 (NEK2) is a microtubule-associated protein that regulates spindle assembly in human cells. It is overexpressed in malignancies. NEK2 upregulation has been observed in primary HCC tissue and HCC cell lines. It is significantly associated with cancer progression, drug resistance, and poor patient prognosis [
21]. In contrast to these findings, another study has reported that low expression of NEK2 is associated with HCC progression and poor patient prognosis [
22].
Lymphokine-activated killer T-cell-originated protein kinase (PBK) is a serine/threonine-protein kinase associated with the dual specific mitogen-activated protein kinase kinase (MAPKK) family. PBK is involved in the activation of lymphoid cells, which support testicular function and spermatogenesis. Overexpression of PBK has been associated with tumor progression. In HCC, PBK overexpression can promote metastasis through activation of the ETV4-uPAR signaling pathway [
23].
Protein regulator of cytokinesis 1 (PRC1) is a protein that regulates antiparallel microtubule cross-linking, promoting the formation of microtubules that support cell shape and regulate cytokinesis. PRC1 is overexpressed in HCC. Its high expression correlates with early HCC recurrence and chemoresistance [
24].
Rac GTPase-activating protein 1 (RACGAP1) belongs to the family of Rho GTPase-activating proteins. It plays a key role in controlling the initiation of cytokinesis, transformation, invasive migration, and metastasis. Overexpression of RacGAP1 protein in colorectal cancer is significantly associated with higher T stage, vessel invasion, and distant metastasis. It is therefore significantly related to poor disease-free and overall survival rates [
25]. In HCC, overexpression of RACGAP1 correlates with early recurrence and a high migration rate [
26].
Ribonucleotide reductase subunit M2 (RRM2) catalyzes the production of deoxynucleotide triphosphates, which are essential for both replicative and repair DNA synthesis. RRM2 is known to play an active role in tumor progression. Elevated RRM2 levels are correlated with poorer prognosis in patients with several types of cancer [
27]. Overexpression of RRM2 in HCC correlates with early recurrence and intrahepatic metastasis. It is therefore associated with poor prognosis [
28].
The DNA topoisomerase 2 α (
TOP2A) gene located on human chromosome 17 encodes the enzyme DNA topoisomerase IIα. TOP2A expression may be a significant predictor of tumor progression, recurrence, and poor survival rate in various types of cancer [
29]. In HCC, high expression of TOP2A has been associated with more advanced clinical stages, lower grades of tumor differentiation, high T stages, and resistance to chemotherapy [
30].
Patterns of expression of the 14 hub genes identified in this study were similar in other malignancies. In addition, several in vivo, in vitro, and bioinformatics studies have shown that individual expression level of each of these 14 hub genes is correlated with the development and progression of HCC.
The background purpose of the present study was to identify key genes and pathways of HBV-associated HCC as a preliminary study before actual experimental study using tissue samples obtained from our own patients. It is essential to perform external validation studies focusing on results of the present study.
The present study has limitations. This study included only one public database. It is necessary to validate results of this study with external data sets obtained from patients of our institution and other Korean institutions.
In conclusion, this comprehensive bioinformatics analysis identified candidate genes and pathways that might be involved in the progression of HBV-associated HCC. These findings may provide better understanding of the molecular mechanisms and potential biomarkers of HCC. Although further experiments with additional patients are required to confirm these findings, results of the present study may suggest future genomic treatment targets for patients with HCC.
Go to :





 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download