I. Introduction
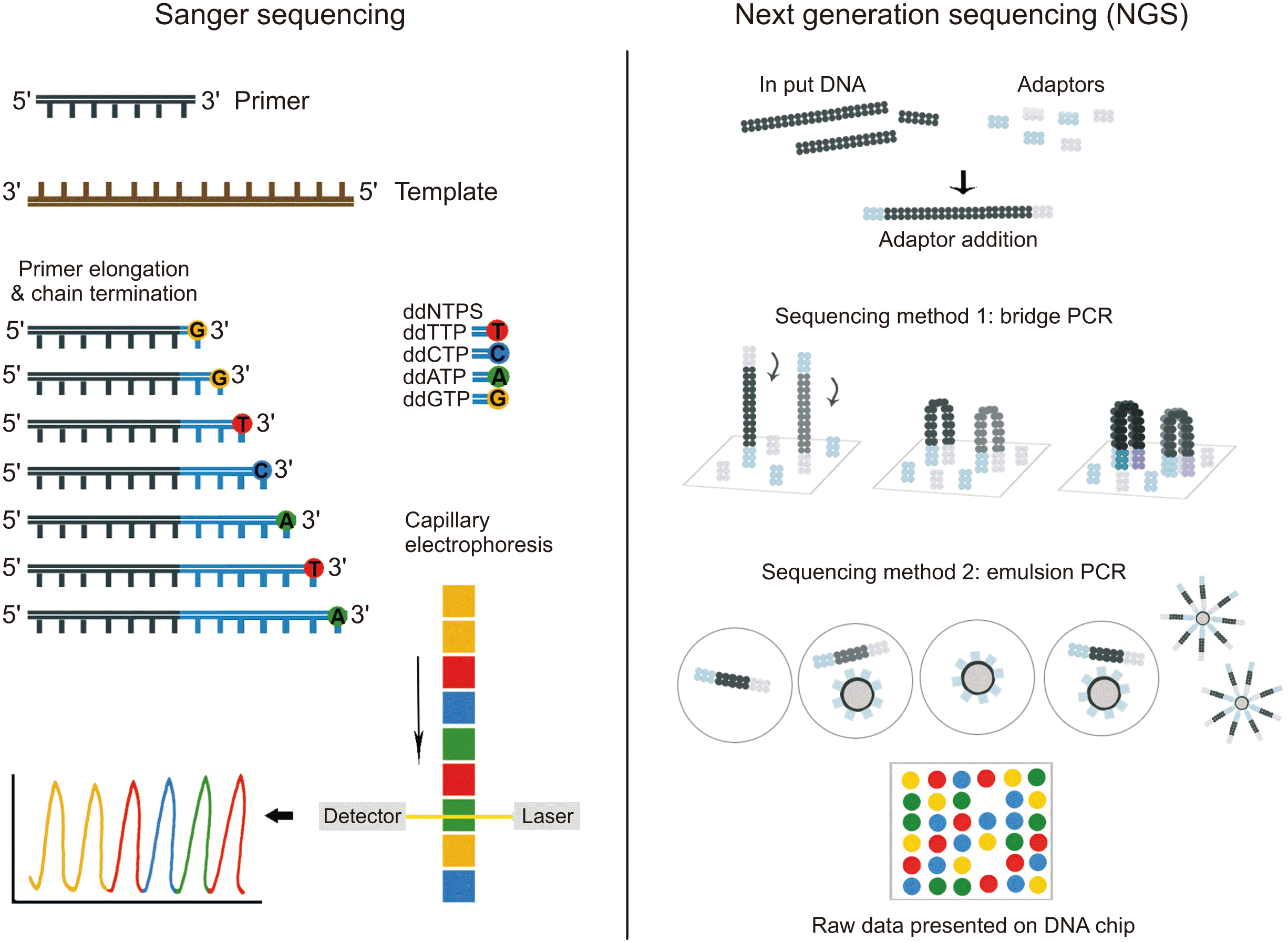
II. Definitions and Use of NGS
Table 1
| Platform | Template preparation | Detection method | NGS coverage (base) | Run time (day) | Gb per run | Essential descriptions |
|---|---|---|---|---|---|---|
| Roche 454 (Roche, Basel, Switzerland)18 | Emulsion-based clonal amplification | Pyrosequencing | 400* | 0.42 | 0.40-0.60 | First commercial platform for the NGS technology. The DNA amplification process is different from that of Illumina, which can sequence much longer reads19. |
| GS FLX Titanium (Roche)20 | 400* | 0.42 | 0.035 | Able to sequence 400-600 million base pairs per run with 400-500 base pair read lengths21. | ||
| Illumina MiSeq (Illumina, San Diego, CA, USA)22 | Clonal bridge PCR | Reversible dye terminator | 2×300 | 0.17-2.7 | 15 |
End-to-end sequencing solutions with reversible- terminator sequencing-by-synthesis. Smallest benchtop sequencer that can perform onboard cluster generation, amplification, genomic DNA sequencing, and data analysis in a single run. Performs both single- and paired-end runs with adjustable read lengths from 1×36 base pairs to 2×300 base pairs22. |
| Illumina HiSeq (Illumina)23 | 2×150 | 0.3-11 | 1,000 |
Generate up to 1,000 Gb per run with the highest yield of data greater than Phred quality score of 30 (Q30). 1 hour’s cycle time can be reduced to 10 minutes23. |
||
| Illumina Genome Analyzer IIX (Illumina)24 | 2×150 | 2-14 | 95 |
Having a broad spectrum of genomic variation with short- and long-insert paired-end reads with insert sizes 200 bp to 5 kb. Used for studying the genome, epigenome, and transcriptome, and also yield greater than 85% of bases higher than Q30 at 2×50 bp24. |
||
| Life Technologies SOLiD4 (Life Technologies, Waltham, MA, USA)25 | Emulsion-based clonal amplification | Oligonucleotide ligation detection | 35-50 | 4-7 | 35-50 | Generates 108-109 small sequence reads at one time, and two-base base encoding to decode the raw data. This system utilizes four fluorescent dyes to interrogate all sixteen (42) possible two-base combinations, by a number of probes. Each probe is eight nucleotides long (8-mer)26. |
| Life Technologies Ion Protons (Thermo Fisher Scientific, Waltham, MA, USA)27 | Native deoxyribonucleotide triphosphates, proton detection | 200 | 0.5 | 100 | Does not use fluorescence or chemiluminescence. Instead, measures the H+ ions released during base incorporation. The lack of any optics allows rapid expansion of the output by approximately 10-fold every six months27. | |
| Complete Genomics (Complete Genomics, San Jose, CA, USA)28 | Gridded DNA-nanoballs | Oligonucleotide ligation detection | 7×10 | 11 | 3,000 |
A DNA nanoball sequencing, which assembles short DNA sequences into a full genome. The sequences are obtained by probe-ligation, but the clonal DNA amplification is performed by rolling circle amplification unlike the bead or emulsion amplification28. |
| Helicos Biosciences Heliscope (Helicos Biosciences, Cambridge, MA, USA)29 | Single molecule | Reversible dye terminator | 35* | 8 | 25 | A highly sensitive fluorescence detection system for direct interrogation of single DNA molecules via sequencing by synthesis30. |
| Pacific Biosciences SMRT (Pacific Biosciences, Menlo Park, CA, USA)31 | Phospholinked fluorescent nucleotides or real-time sequencing | 10,000 (N50); 30,000+ (max) | 0.08 | 0.5 |
Long-read sequencing platforms with SMRT sequencing technology. Template preparation does not require any amplification steps, and the prepared library molecule is the sequencing template. The adapters have a hairpin structure (SMRT loop adapters) so that after ligation the double stranded DNA fragments will have become circular31. |
III. Salivary Biomarkers and Oral Cancer
Table 2
(DAPK: death-associated protein kinase, TIMP3: tissue inhibitor of metalloproteinase-3, MGMT: O6-methylguanine-DNA methyltransferase, Maspin: mammary serine protease inhibitor, IL-8: interleukin 8, IL-1β: interleukin 1 beta, S100P: S100 calcium binding protein P, SAT: spermidine/spermineN1-acetyltransferase, miR: microRNA, CD44: cluster of differentiation 44, 8-OHdG: 8-Oxo-2'-deoxyguanosine, ANGPT: angiopoietin, VEGF: vascular endothelial growth factor, HPV: human papilloma virus, EBV: Epstein–Barr virus)
IV. Current Cancer Biomarkers Using NGS-based Technique
Table 3
| Potential genomic markers1 | Sample/ Collection method | NGS-based studies |
|---|---|---|
| p53 (TP53, P53; BCC7; LFS1; BMFS5; TRP53)46,47 | Tumor tissue/ Formalin fixation and paraffin embedding | Approximately 30% of salivary gland cancers have mutations of p53 gene. A significant worse overall and disease-free survival was shown in cancers with p53 mutations. |
| Promoter hypermethylation of DAPK gene48,49 |
UWS/ Oragene DNA Self-Collection kit Salivary rinse/ 10-20 mL 0.9% NaCl, 15-60 s |
DAPK methylation gene was reported with the development of cancers in women. |
| Promoter hypermethylation of TIMP3 gene (SFD; K222; K222TA2; HSMRK222)50-52 | Salivary rinse/ rinsing and gargling with 20 mL 0.9% NaCl | Promoter hypermethylation of TIMP3 levels found in the circulating tumor DNA in saliva of disease-free survival HNSCC patients. |
| Promoter hypermethylation of p16 (CDKN2A, ARF; MLM; P14; P19; CMM2; INK4)47,53 |
Tumor tissue/ Formalin fixation and paraffin embedding Salivary rinse/ swishing with 15 mL of 0.9% normal saline, 15-30 s |
The absolute frequencies of CDKN2A mutations are detected by NGS. |
| Promoter hypermethylation of MGMT gene54 |
Tumor tissue/ Formalin fixation and paraffin embedding Salivary rinse/ swishing with 15 mL of 0.9% normal saline, 15-30 s |
MGMT methylation was detected in 29% of oral cancer/ dysplasia patient, showing MGMT gene is related to DNA repair and aberrant promoter hypermethylation. |
| Cyclin D1 gene amplification (CCND1, BCL1; PRAD1; U21B31; D11S287E)55,56 | Tumor tissue/ Formalin fixation and paraffin embedding | 14 of the 21 genes had copy number amplifications and losses, which included CCND1 (83.7%). |
| Maspin57,58 | UWS/ samples were collected in the morning, without any oral stimulation for 90 minutes before collection | Downregulation of maspin; the functional importance of maspin includes the inhibition of tumor angiogenesis. The levels of maspin were reduced. |
Table 4
| Potential mRNA markers1 | Sample/ Collection method | NGS-based studies |
|---|---|---|
| IL-8 (IL8)59,60 | UWS/ samples were collected between 9 am and 10 am, following the standard protocol. | In oral, esophageal, lung, pancreatic, ovarian and breast cancers, certain salivary mRNA biomarkers have been proposed as a possible cancer biomarker, including IL-8. |
| IL-1β (IL-1)61,62 | UWS/ 3-5 mL salivary specimen was collected into a tube containing 10 mL of RNAlater (Ambion, Austin, TX, USA), an aqueous tissue storage reagent that rapidly permeated tissue to stabilize and protect cellular RNA. The specimen was then placed on ice at 4°C; cell-free saliva supernatant was harvested. | IL-1β gene has been found to be an important biomarker for ovarian cancer. |
| S100P (S100 calcium binding protein P)63,64 | UWS/ samples were collected between 6 am and 12 pm following standard protocol. A maximum of 8 mL of saliva were collected within 30 minutes. | Salivary S100 mRNA is a candidate biomarker for detecting OSCC development and in OLP patients determined by NGS. |
| SAT69 | UWS/ Participants were asked to refrain from eating, drinking and any oral hygiene overnight and spit in 5-mL plastic vials used for biochemical examinations for 5 minutes. During the whole procedure and until centrifugation the vials were kept in ice. | The combination of SAT and IL-8 mRNA biomarkers are attractive candidates either for screening or for early diagnosis purposes. These exert a very good prediction ability together with a high sensitivity and specificity for screening oral squamous cell carcinoma. |
| miR 31 (MIRN31; miR-31; hsa-mir-31)65,66 | UWS/ 3-5 mL saliva was collected from mouth floor after simple rinsing. Pre-treatment salivary sample was collected from 45 patients with OSCC and 10 patients with oral verrucous leukoplakia and 24 healthy participants. | A significantly high expression of miR-31 was found in the saliva sample of patients with OSCC at all clinical stages by RT-qPCR. More miR-31 levels were detected in the saliva than in plasma, suggesting salivary miR-31 to be a more sensitive. |
| miR 125 (MIRN125A; mir-125a; miRNA125A)36,67 | UWS/ an aqueous tissue storage reagent was used to preserve UWS samples and SUPERase.In (Themo Fisher Scientific, Waltham, MA, USA) was used for supernatant saliva preservation. | Significantly lower levels of miR-125a was found in saliva sample of OSCC patient than that of healthy controls. |
| miR 200a36,68 | UWS/ an aqueous tissue storage reagent was used for the UWS samples and SUPERase.In (Themo Fisher Scientific) was used for supernatant saliva preservation. | Significantly lower levels of miR 200a was found in saliva sample of OSCC patient than that of healthy controls. |
Table 5
| Genomic | Transcriptome (mRNA) | |
|---|---|---|
| Known salivary biomarkers |
p53 FAT1 CASP8 PIK3CA HRAS NOTCH1 CDKN2A70 |
miRNA (saliva): miR-let-7a-5p and miR-392874 Oncogenic (up-regulated, tissue): miR-21, miR-22, miR-26a, miR34c, miR-34b, miR-117, miR-118, miR-130b, miR-135, miR-142, miR-143, miR-148a, miR-150, miR-221, miR-222, miR-423, miR-542, miR-1269a81 Suppressive (down-regulated, tissue): miR-92b, miR-199, miR-214, miR-375, miR-486, miR-504, miR-499, miR-48681 |
| Recommended NGS-based salivary biomarker |
Mutation of p53 gene Promoter hypermethylation of DAPK, TIMP3, p16, and MGMT genes Cyclin D1 gene amplification Mammary serine protease inhibitor |
miR 3176, miR 12577, miR 20078,79 |
(NGS: next generation sequencing, FAT1: AT atypical cadherin 1, CASPS: caspase 8, PIK3CA: phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha, HRAS: Harvey rat sarcoma viral oncogene homolog, NOTCH1: Notch homolog 1, translocation-associated, CDKN2A: cyclin-dependent kinase inhibitor 2A, DAPK: death-associated protein kinase, TIMP3: tissue inhibitor of metalloproteinase-3, MGMT: O6-methylguanine-DNA methyltransferase, miR: microRNA)




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download