Introduction
Materials and methods
1. Reagents
2. Procurement of rheumatoid arthritis synovial tissues and immunofluorescence staining
3. Human peripheral blood mononuclear cells preparation
4. Mouse peritoneal macrophage preparation
5. Sulf-1, Sulf-2 knockdown
6. Sulf-1, Sulf-2 overexpression
7. Antigen presentation assay
8. Phagocytosis assay
9. Real-time polymerase chain reaction
10. Western blotting
11. Statistical analyses
Results
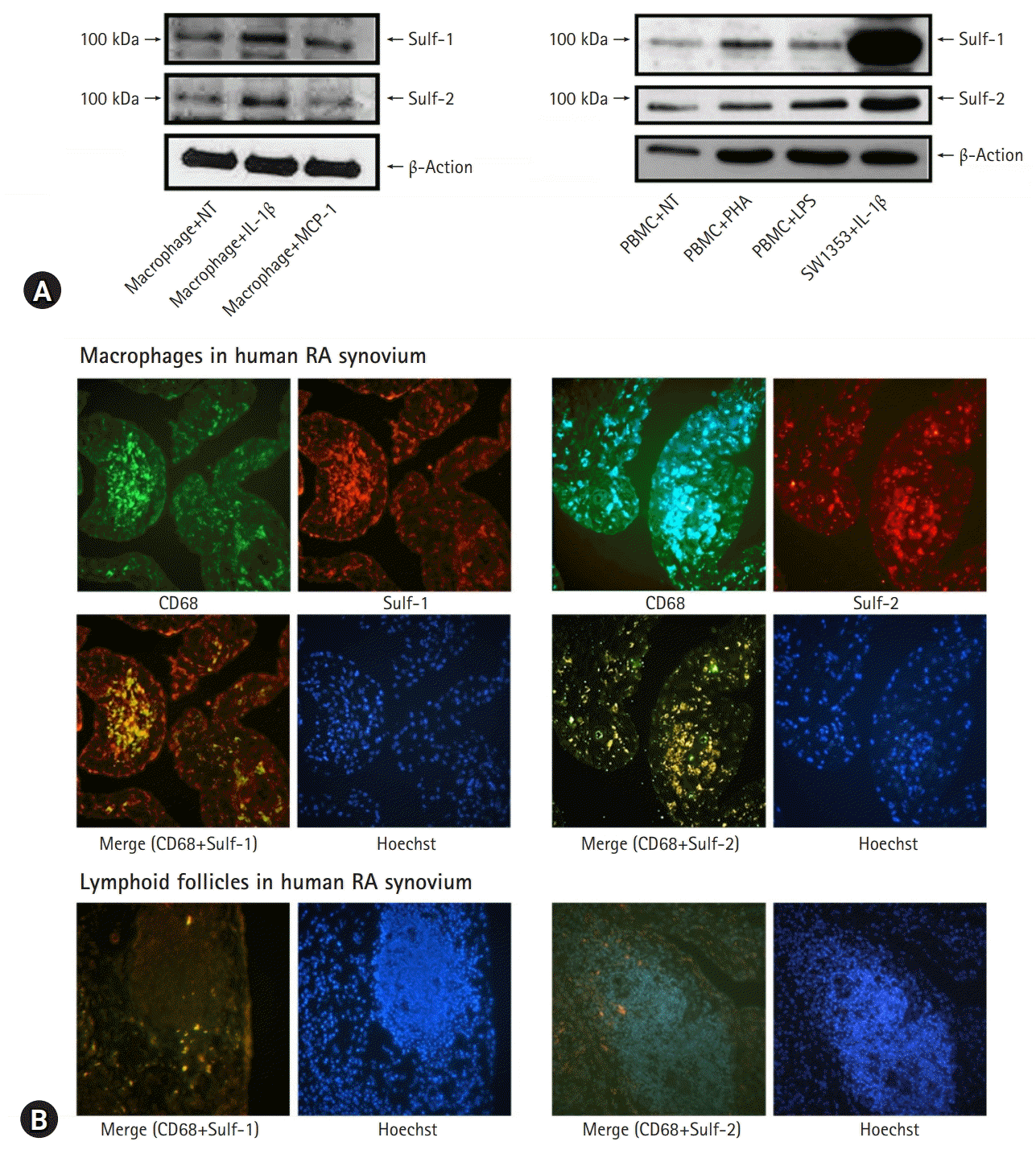 | Fig. 1.Sulfatase (Sulf) expression in the inflammatory cells with proinflammatory stimuli and tissue inflammation. Murine peritoneal macrophages show increased expression of Sulf-1 and Sulf-2 upon stimulation with interleukin (IL)-1β. (A) However, normal non-treated (NT) peripheral blood mononuclear cells (PBMCs) show weak expression of Sulfs and no change in Sulf expression by in vitro activation with phytohemagglutinin and lipopolysaccharides (LPS) (SW1353, a cell line, as positive control) (two-colored double immunofluorescence stain, x200). (B) Rheumatoid arthritis (RA) synovial tissues show chronic inflammatory reactions. Synovial macrophages as CD68 positive cells infiltrated in sublining layers. Lymphocytes as the CD68 negative cells of small nuclei clustered in lymphoid follicles. (B, upper panel) Two-colored double immunofluorescence staining of Sulfs and CD68 revealed synovial macrophages colored bright yellow as the result of dual positive staining of CD68 and Sulfs. (B, lower panel) However, lymphocytes are completely negative for both CD68 and Sulfs (two-colored double immunofluorescence stain, x200). MCP-1, monocyte chemotactic protein-1. |
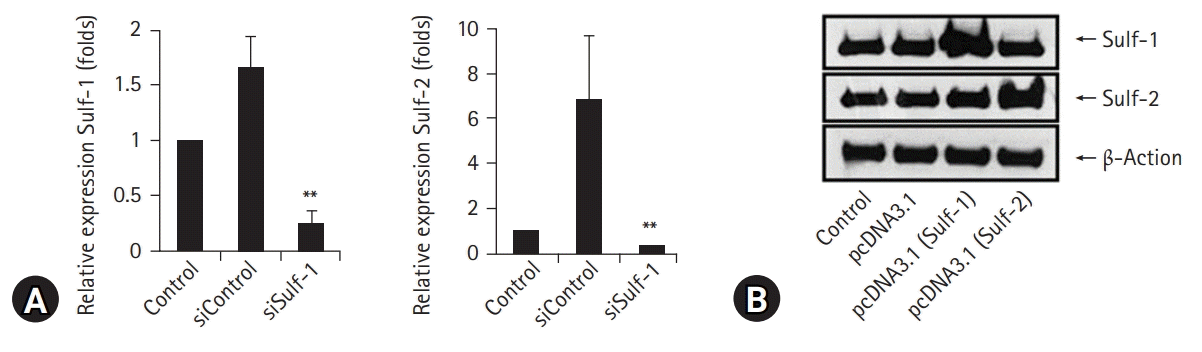 | Fig. 2.Macrophage sulfatase (Sulf) expression was knocked down using small interfering (si) RNAs against Sulf-1 and Sulf-2, and upregulated by transfection with pcDNA3.1/Sulf-1 and pcDNA3.1/Sulf-2 plasmids. (A) Downregulation of Sulf gene transcription in macrophages transfected with Sulf-1 and Sulf-2 siRNAs, as compared with non-treated (NT) and siRNAs controls, was confirmed by real-time polymerase chain reaction. (B) Overexpression of Sulf-1 and Sulf-2 in macrophages transfected with pcDNA3.1/Sulf-1 and Sulf-2 plasmids, as compared with those of NT and pcDNA3.1 controls, was confirmed by western blot. **p<0.01. |
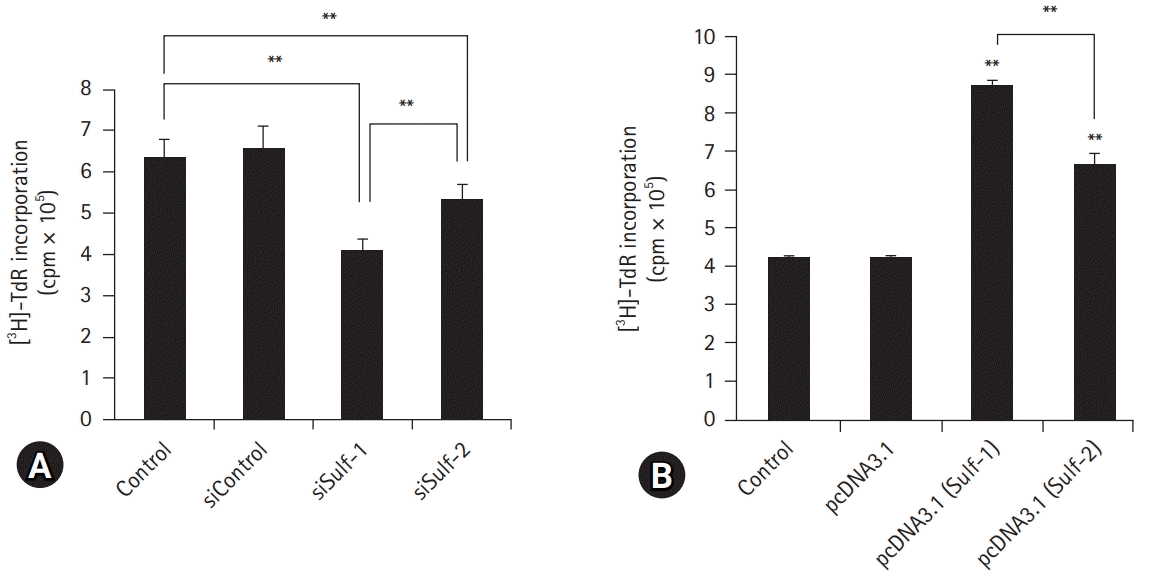 | Fig. 3.Antigen presentation assay with modification of sulfatase (Sulf) expression. Murine peritoneal macrophages were cultured with ovalbumin. (A) Macrophages with Sulf-1 or Sulf-2 suppression show significantly lower [3H]-thymidine radioactivity incorporated into interleukin-2 dependent CTLL-2 cells as compared with controls. (B) Macrophages with upregulation of Sulf-1 or Sulf-2 show significantly higher [3H]-thymidine radioactivity compared with controls. The [3H]-thymidine radioactivity is reciprocal by Sulf suppression and overexpression. The changes in radioactivity are more prominent with Sulf-1 regulation than with Sulf-2. cpm, counts per minute. **p<0.01. |
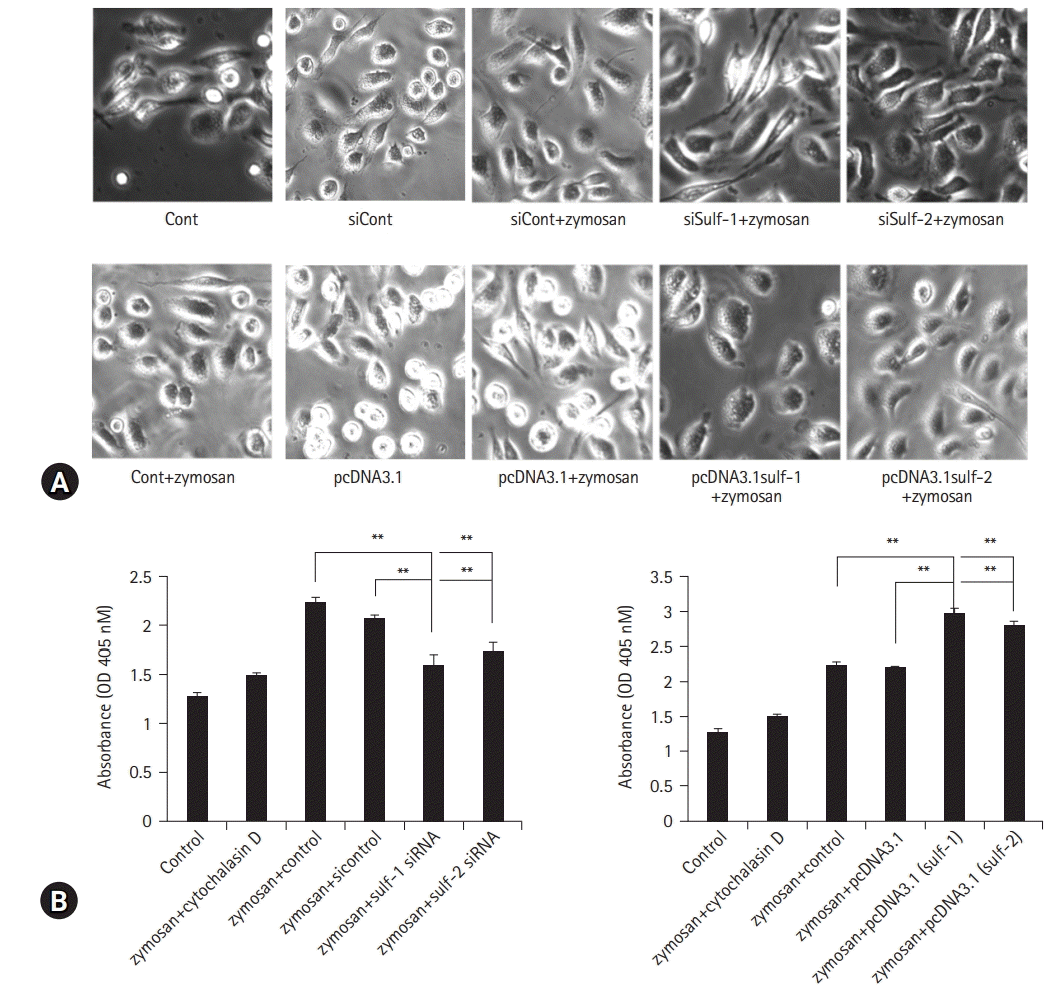 | Fig. 4.Macrophage phagocytosis assay with modification of sulfatase (Sulf) expression. Macrophages were cultured with zymosan. (A, upper row) Sulfs-knockdown macrophages show marked suppression of zymosan phagocytosis compared with controls (Cont) on microscopic images. (A, lower row) Sulfs-overexpressing macrophages show brisk phagocytotic activity against zymosan. The phagocytic activity was analyzed by measuring the absorbance at 450 nm using spectrophotometry. (B, left) The absorbance of Sulfs-knockdown macrophages is significantly lower than that of controls. (B, right) Sulfs-overexpressing macrophages demonstrated higher absorbance compared with controls. The effects of Sulfs on macrophage phagocytosis are more prominent with Sulf-1 than with Sulf-2. si, small interfering; OD, optical density; AU, absorbance unit. **p<0.01. |




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download