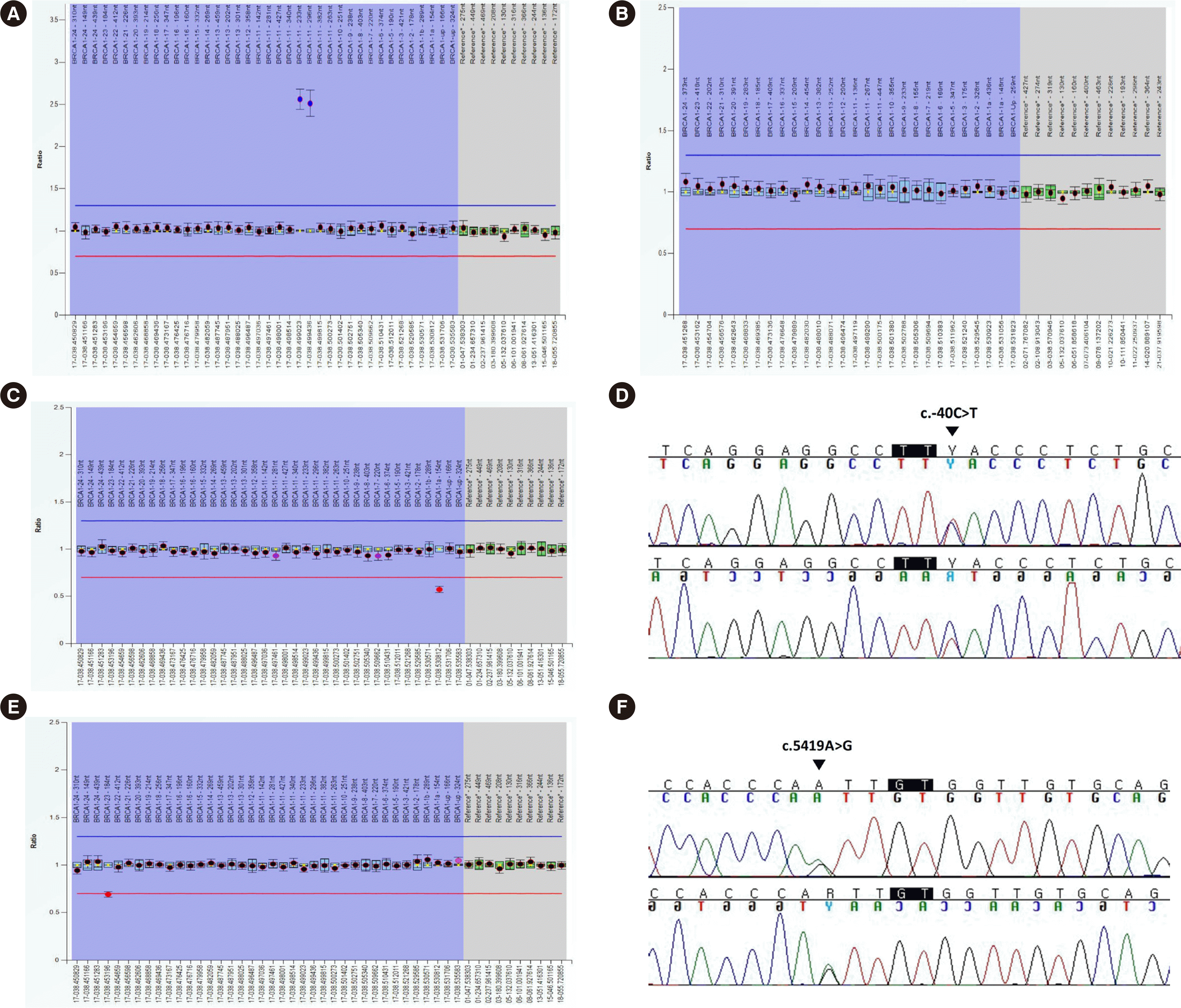
 | Fig. 1Results of initial MLPA and confirmative tests of the three cases. (A) In the first case, an initial MLPA test with the P002 kit showed abnormal signals, suggesting partial duplication of exon 11 of BRCA1. (B) However, a subsequent confirmative MLPA test using P087 showed no abnormal results. An additional P002 MLPA test using a new venous blood sample showed all signals within normal limits. (C) In the second case, an initial P002 MLPA test revealed a decreased DQ 0.57 for the probe set for exon 1a, suggesting heterozygous deletion of this exon. (D) Direct sequencing of BRCA1 exon 1a revealed a single nucleotide substitution c.-40C>T (indicated by the black arrowhead) in the two-nucleotide region flanking the probe hybridization site. (E) In the third case, an initial P002 MLPA test revealed a decreased DQ 0.69 for the probe set for exon 23. (F) Direct sequencing of exon 23 revealed c.5419A>G (indicated by the black arrowhead) in the four-nucleotide region flanking the probe hybridization site. Probe hybridization sites are indicated in black rectangles.
Abbreviations: DQ, dosage quotient; MLPA, multiplex ligation-dependent probe amplification.
|
Dear Editor,
BRCA1 and BRCA2 are the major genes involved in hereditary breast and ovarian cancer [1]. Large genomic rearrangements (LGRs) have been reported to be pathogenic variants in BRCA1/2 [2, 3]. Various molecular diagnostic methods, such as next-generation sequencing (NGS) and digital droplet PCR, have been used to detect LGRs in BRCA 1/2 [4–6]. However, these methods have not been sufficiently validated for clinical diagnostic purposes and make use of platform instruments that are expensive and require well-trained, expert personnel. Therefore, multiplex ligation-dependent probe amplification (MLPA) has been considered an efficient alternative for identifying LGRs. MLPA has disadvantages, such as false-positive results due to alterations in the probe-binding or ligation site sequences [7–10]. Here, we present three cases of false-positive BRCA1 MLPA test results. This study was approved by the Institutional Review Board (IRB) of Keimyung university Dongsan hospital, Daegu, Korea (IRB No. 2021-08-074-003). Informed consent for reporting the cases was waived since the study involved retrospective review of medical records. However, informed consent was obtained for clinical diagnostic DNA testing.
A 52-year-old female patient diagnosed with breast cancer underwent a BRCA 1/2 test at Keimyung University Dongsan Hospital in Nov. 2020. DNA was extracted from peripheral blood leukocytes. Whole exons of BRCA1/2 were analyzed by NGS using the Oncomine BRCA Research Assay (Thermo Fisher Scientific, Rockford, IL, USA), which revealed no pathogenic variants or LGRs. MLPA was performed using SALSA MLPA Probemixes P002-D1 BRCA1 and P045-D1 BRCA2 (MRC Holland, Amsterdam, The Netherlands) and an ABI 3500XL genetic analyzer (Applied Biosystems, Foster City, CA, USA). The data were analyzed using Coffalyser.Net software (MRC Holland). Initially, two of the eight probes targeting exon 11 of BRCA1 yielded abnormal dosage quotients (DQs) when compared with that by the control probe (2.56 and 2.6) (Fig. 1A). As these ratios were >1.2, which is the manufacturer’s cut-off for normal DQ, they suggested partial duplication of exon 11. The test was repeated with P002 by another technician, which yielded similar results (DQs, 2.56 and 2.51). An additional BRCA1 MLPA test with P087 did not show an abnormal DQ. A P002 MLPA test was performed with DNA newly extracted from another whole blood sample. The result showed normal DQs across all probes (Fig. 1B).
The second case was of a 55-year-old female patient with ovarian cancer. The BRCA1/2 exons that were sequenced by NGS in Nov. 2020 revealed no pathogenic variants or LGRs. The P002 MLPA test suggested a heterozygous deletion of exon 1a of BRCA1; the DQ was 0.57, which was below the 0.65 cut-off value reported by the manufacturer (Fig. 1C). However, the P087 MLPA test did not show an abnormal DQ, and direct sequencing revealed a single nucleotide substitution in the two-nucleotide region (NM_007294.4:c.-40C>T) flanking the probe hybridization site of exon 1a (probe No. 00763-L22990) (Fig. 1D).
The third case was of a 40-year-old female patient who was tested for BRCA1/2 because of breast cancer. The MLPA test revealed a suspicious heterozygous deletion of exon 23 of BRCA1 (DQ, 0.69) (Fig. 1E). However, NGS and direct sequencing revealed a single nucleotide substitution (NM_007294.4:c.5419A>G) in a four-nucleotide region flanking the probe hybridization site (probe No. 20023-L23035), which was a variant of uncertain significance (Fig. 1F). The P087 MLPA test showed no LGRs.
MLPA is sensitive, inexpensive, and relatively simple, compared to other methods; however, several cases of false-positive results have been reported [8–10]. In the first case, the false-positive duplication was resolved using DNA extracted from another whole blood sample. We believe that the false-positive result was due to contamination of the first DNA extract. In such cases, using other methods, such as sequencing, would not correct the erroneous result. Additional MLPA tests using another kit and resampling must be considered for confirmation. In the other two cases, a deletion was falsely detected in a single exon with one probe set but was not detected by additional MLPA tests. Subsequent sequencing identified the cause of the false test result as a hybridization site mutation, a known cause of false-positive deletions in MLPA tests [9, 10].
These cases and results of previous studies highlight the need for confirmation of duplications or deletions involving single or multiple exons. Direct sequencing of the suspected sites, the use of additional MLPA kits and resampling should be considered to confirm the test results in such cases. Using latest MLPA probes and reagents is also important, as manufacturers continuously develop new reagents to avoid hybridization interference by point mutations. Errors can still occur when using the latest MLPA kits, and results that are clearly below the cut-off can be mistaken as deletions. Thus, in case of suspicious results, confirmatory MLPA tests or direct sequencing should be performed.
Go to : 
Notes
AUTHOR CONTRIBUTIONS
Kim KB and Kim D collected and summarized literature reports, interpreted the test results, and contributed to manuscript writing. Park S participated in interpreting and describing the test results. Ha JS and Ryoo N collected and summarized literature reports and contributed to manuscript writing.
Go to : 
REFERENCES
1. King MC, Marks JH, Mandell JB. New York Breast Cancer Study Group. 2003; Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 302:643–6. DOI: 10.1126/science.1088759. PMID: 14576434.

2. Ewald IP, Ribeiro PL, Palmero EI, Cossio SL, Giugliani R, Ashton-Prolla P. 2009; Genomic rearrangements in BRCA1 and BRCA2: a literature review. Genet Mol Biol. 32:437–46. DOI: 10.1590/S1415-47572009005000049. PMID: 21637503. PMCID: PMC3036053.
3. Hansen Tv, Jønson L, Albrechtsen A, Andersen MK, Ejlertsen B, Nielsen FC. 2009; Large BRCA1 and BRCA2 genomic rearrangements in Danish high risk breast-ovarian cancer families. Breast Cancer Res Treat. 115:315–23. DOI: 10.1007/s10549-008-0088-0. PMID: 18546071.

4. Oscorbin I, Kechin A, Boyarskikh U, Filipenko M. 2019; Multiplex ddPCR assay for screening copy number variations in BRCA1 gene. Breast Cancer Res Treat. 178:545–55. DOI: 10.1007/s10549-019-05425-3. PMID: 31482362.

5. Han E, Yoo J, Chae H, Lee S, Kim DH, Kim KJ, et al. 2020; Detection of BRCA1/2 large genomic rearrangement including BRCA1 promoter-region deletions using next-generation sequencing. Clin Chim Acta. 505:49–54. DOI: 10.1016/j.cca.2020.02.023. PMID: 32092317.

6. Yoo J, Lee GD, Kim JH, Lee SN, Chae H, Han E, et al. 2020; Clinical validity of next-generation sequencing multi-gene panel testing for detecting pathogenic variants in patients with hereditary breast-ovarian cancer syndrome. Ann Lab Med. 40:148–54. DOI: 10.3343/alm.2020.40.2.148. PMID: 31650731. PMCID: PMC6822011.

7. Varga RE, Mumtaz R, Jahic A, Rudenskaya GE, Sánchez-Ferrero E, Auer-Grumbach M, et al. 2012; MLPA-based evidence for sequence gain: pitfalls in confirmation and necessity for exclusion of false positives. Anal Biochem. 421:799–801. DOI: 10.1016/j.ab.2011.12.002. PMID: 22222296.

8. Santos R, Gonçalves A, Oliveira J, Vieira E, Vieira JP, Evangelista T, et al. 2014; New variants, challenges and pitfalls in DMD genotyping: implications in diagnosis, prognosis and therapy. J Hum Genet. 59:454–64. DOI: 10.1038/jhg.2014.54. PMID: 25007885.

9. Maistro S, Teixeira N, Encinas G, Katayama ML, Niewiadonski VD, Cabral LG, et al. 2016; Germline mutations in BRCA1 and BRCA2 in epithelial ovarian cancer patients in Brazil. BMC Cancer. 16:934. DOI: 10.1186/s12885-016-2966-x. PMID: 27914478. PMCID: PMC5135756.
10. Kim MJ, Cho SI, Chae JH, Lim BC, Lee JS, Lee SJ, et al. 2016; Pitfalls of multiple ligation-dependent probe amplifications in detecting DMD exon deletions or duplications. J Mol Diagn. 18:253–9. DOI: 10.1016/j.jmoldx.2015.11.002. PMID: 26743743.
Go to : 




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download