Dear Editor,
Clostridium saudiense (formerly known as C. saudii) is a spore-forming, anaerobic, gram-positive bacillus, first identified by Angelakis, et al. [1] from the stool of an obese 24-year-old Saudi individual and named C. saudii. As C. saudii was not a validly published name, the bacterium was later renamed C. saudiense [2]. Extensive culturomics studies of stool samples from healthy individuals have indicated that C. saudiense is a commensal bacterium colonizing the human intestinal tract [3-5]. To date, no apparent infections caused by C. saudiense have been reported. We report the first case of C. saudiense bacteremia. The Institutional Review Board (IRB) of Samsung Medical Center, Seoul, Korea (IRB number SMC 2021-08-077), approved this study and waived the need for informed consent as this study poses minimum risk to the patient.
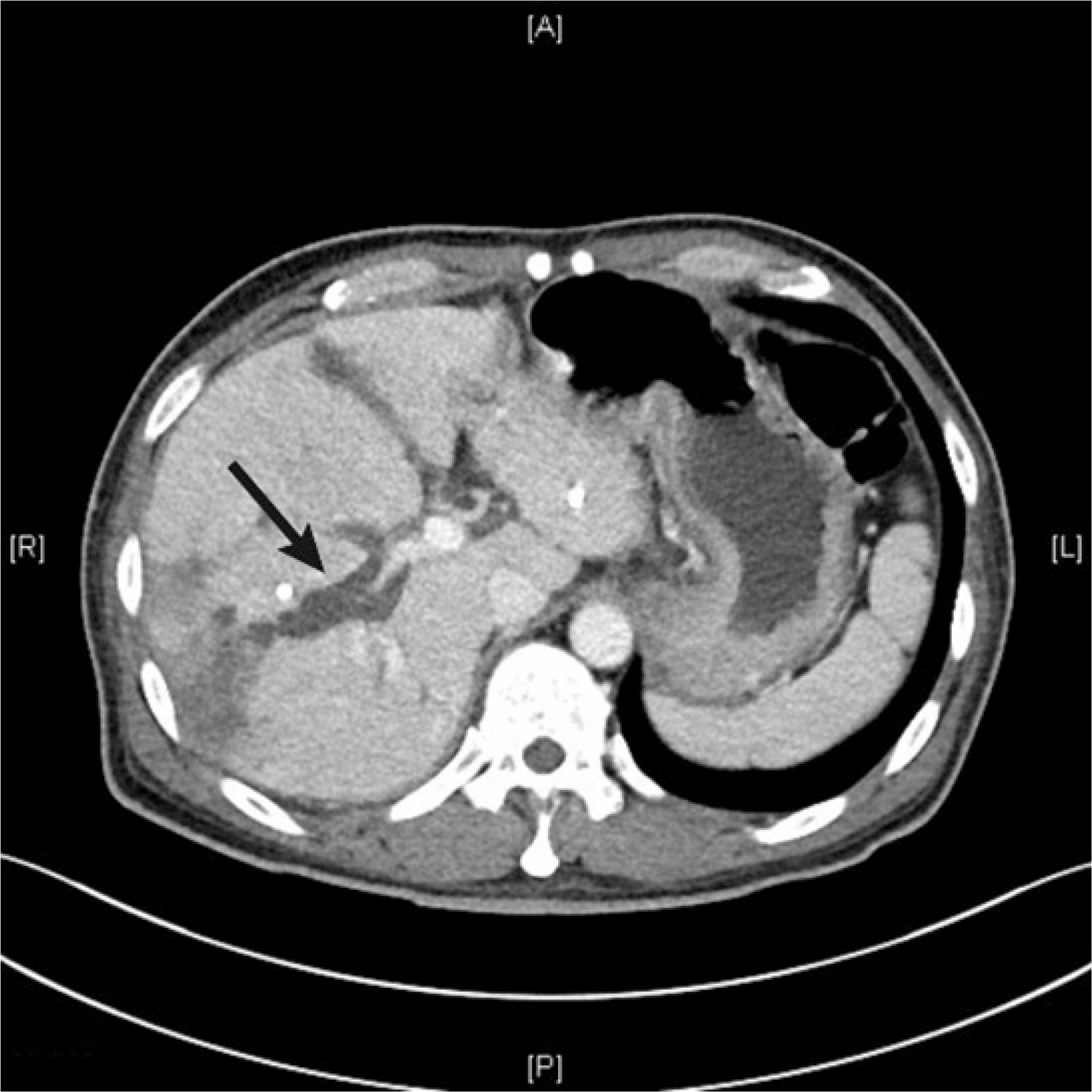
A 49-year-old male patient with hepatocellular carcinoma visited the Samsung Medical Center emergency room complaining of persistent fever and abdominal discomfort. He had undergone a transcatheter arterial chemoembolization (TACE) procedure one month prior. His temperature was 38.4°C, blood pressure 98/65 mmHg, pulse rate 99 beats/min, and respiratory rate 20 breaths/min. Laboratory parameters were as follows: leukocyte count, 14.76×109/L (neutrophils, 78.8%) and C-reactive protein (CRP) level, 150.5 mg/L (reference interval, 0.0–5.0 mg/L). Abdominopelvic computed tomography revealed ischemic biliopathy and intrahepatic retention of bile, forming a biloma (Fig. 1).
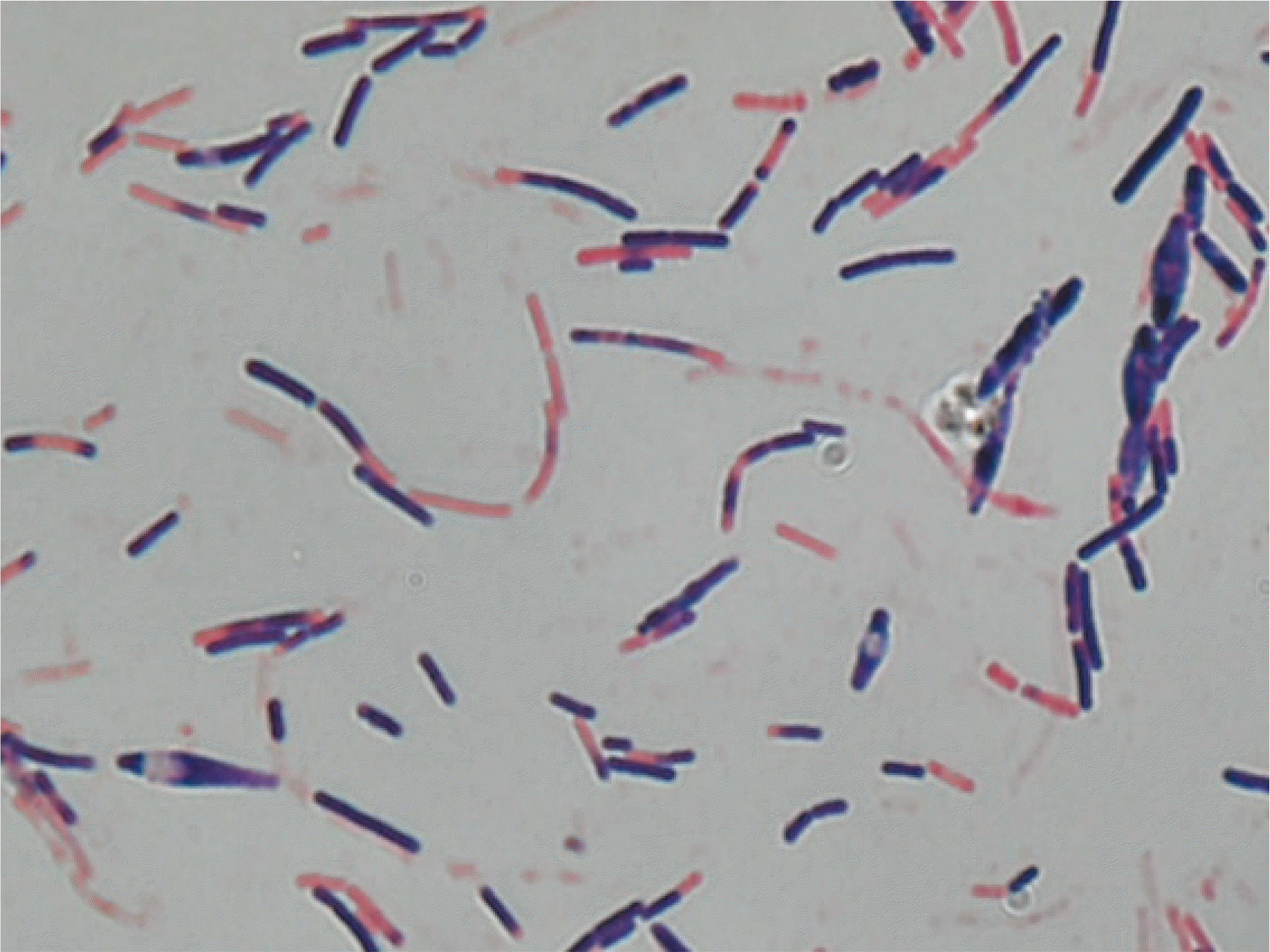
Two sets of blood samples from peripheral veins were collected in BACT/ALERT FA/FN PLUS bottles (bioMérieux, Marcy-l’Étoile, France) and incubated in the BACT/ALERT VIRTUO system (bioMérieux). The patient was subjected to empirical antibiotic therapy with ceftriaxone 2,000 mg intravenously (IV) every 24 hours and metronidazole IV 500 mg every 8 hours on day 1. The next day, the antibiotic was switched to piperacillin/tazobactam IV 3,375 mg every 6 hours owing to persistent fever. On day 3 of incubation, positive growth was observed in one anaerobic bottle of the two sets of blood cultures. Gram staining revealed gram-variable, rod-shaped bacteria of various lengths and occasional endospores (Fig. 2). The positive blood culture was plated onto blood agar, MacConkey agar, and Brucella agar for sub-culture. Growth was observed only on Brucella agar under anaerobic conditions. Bacterial identification using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (VITEK MS; bioMérieux) was unsuccessful. The VITEK 2 system using the ANC card (bioMérieux) identified the isolate as Clostridium clostridioforme with a probability of 88%, failing precise identification.
For accurate identification, we performed 16S ribosomal RNA gene sequence analysis as described previously [6]. The sequence obtained was compared with those in the GenBank database using the basic local alignment search tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The sequence matched that of C. saudiense JCC (GenBank accession number NR_144696.1) perfectly (100.0%, 1,411/1,411 bp). The second-best match was Clostridium celatum, with 98.9% sequence identity (1,397/1,412 bp). Based on these findings, the isolate was identified as C. saudiense [7].
The patient was treated with piperacillin/tazobactam IV 3,375 mg every 6 hours for two weeks followed by an additional four weeks of oral metronidazole 250 mg every 8 hours and oral cefixime 300 mg every 24 hours. C. saudiense was re-cultured from bile samples obtained via percutaneous drainage and two sets of additional blood cultures on day 3. Follow-up blood cultures on days 5 and 7 were negative, and the patient’s CRP level started to decline. After scheduled antibiotic therapy, the patient showed no symptoms or signs of infection and the CRP level normalized.
Clostridium species are widespread in nature, and some are present in the human intestinal tract, female genital tract, and oral mucosa as commensals [8]. Clostridium species are the second most common anaerobes isolated from blood [9], especially in patients with predisposing factors, such as gastrointestinal anomalies or immunocompromise [8, 10]. However, C. saudiense infections, particularly bacteremia, have not been reported. In our case, commensal C. saudiense translocated from the gastrointestinal system to biliary system led to a loculated biloma and eventually developed into a bloodstream infection. Like in other cases of clostridial bacteremia, the patient had a predisposing factor, namely a recent intervention history (TACE). The patient responded well to empirical treatment for Clostridium species.
In conclusion, we report the first case of C. saudiense bacteremia. Our findings suggest that C. saudiense be considered an opportunistic pathogen with the potential to cause biliary infection and bacteremia in patients with damage to the biliary system.
Go to : 
Notes
AUTHOR CONTRIBUTIONS
Huh HJ designed the study. Heo WY, Kang OK, Kim TY, and Yu HJ analyzed the data. Yoon EJ drafted the manuscript. Lee JH, Ko JH, Huh HJ, and Lee NY reviewed the manuscript. Submission of the final manuscript was approved by all authors.
Go to : 
REFERENCES
1. Angelakis E, Bibi F, Ramasamy D, Azhar EI, Jiman-Fatani AA, Aboushoushah SM, et al. 2014; Non-contiguous finished genome sequence and description of Clostridium saudii sp. nov. Stand Genomic Sci. 9:8. DOI: 10.1186/1944-3277-9-8. PMID: 25780501. PMCID: PMC4334108.
2. Oren A, Garrity GM. 2016; List of new names and new combinations previously effectively, but not validly, published. Int J Syst Evol Microbiol. 66:2463–6. DOI: 10.1099/ijsem.0.001149. PMID: 27530111.

3. Diakite A, Dubourg G, Dione N, Afouda P, Bellali S, Ngom II, et al. 2019; Extensive culturomics of 8 healthy samples enhances metagenomics efficiency. PLoS One. 14:e0223543. DOI: 10.1371/journal.pone.0223543. PMID: 31634343. PMCID: PMC6802823.

4. Lagier JC, Hugon P, Khelaifia S, Fournier PE, La Scola B, Raoult D. 2015; The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 28:237–64. DOI: 10.1128/CMR.00014-14. PMID: 25567229. PMCID: PMC4284300.

5. Dubourg G, Baron S, Cadoret F, Couderc C, Fournier PE, Lagier JC, et al. 2018; From culturomics to clinical microbiology and forward. Emerg Infect Dis. 24:1683–90. DOI: 10.3201/eid2409.170995. PMID: 30124405. PMCID: PMC6106433.

6. Park JH, Kim TS, Park H. 2020; A Hungatella effluvii isolate in blood culture of a patient with hematochezia. Anaerobe. 63:102211. DOI: 10.1016/j.anaerobe.2020.102211. PMID: 32417531.
7. CLSI. 2018. Interpretive criteria for identification of bacteria and fungi by targeted DNA sequencing. Clinical Laboratory Standards Institute;Wayne, PA: 2nd ed. CLSI MM18.
8. Jorgensen J, Pfaller M, editors. 2015. Manual of clinical microbiology. 11th ed. American Society of Microbiology;Washington DC: p. 940–66.
9. Simmon KE, Mirrett S, Reller LB, Petti CA. 2008; Genotypic diversity of anaerobic isolates from bloodstream infections. J Clin Microbiol. 46:1596–601. DOI: 10.1128/JCM.02469-07. PMID: 18322067. PMCID: PMC2395081.

10. Brook I. 2010; The role of anaerobic bacteria in bacteremia. Anaerobe. 16:183–9. DOI: 10.1016/j.anaerobe.2009.12.001. PMID: 20025984.

Go to : 




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download