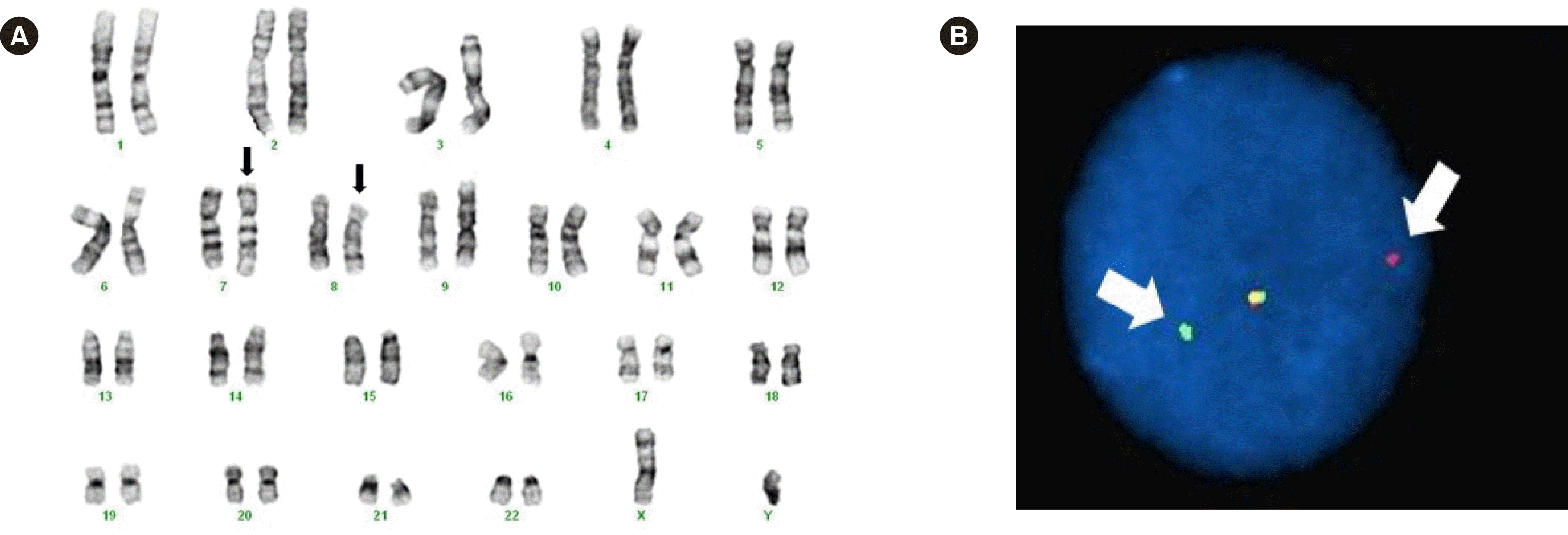
 | Fig. 2Cytogenetic analysis of a bone marrow specimen. (A) G-banded karyogram of bone marrow cells revealing 46,XY,t(7;8)(q34;p11.2), i(9)(q10). The arrows indicate the chromosomes involved in this translocation. (B) FISH analysis using an FGFR1 break-apart probe (XL FGFR1 probe; MetaSystems, Altlussheim, Germany). The arrows indicate split signals related to FGFR1 rearrangement. |
Dear Editor,
According to the 2016 WHO classification, myeloid/lymphoid neoplasms associated with clonal eosinophilic disorders are associated with specific genes, such as PDGFRA, PDGFRB, FGFR1, and PCM1-JAK2 [1]. FGFR1 rearrangement has been reported in myeloid/lymphoid neoplasms of various disease groups and associated with various partner genes [1, 2]. We report a rare case of B-lymphoblastic leukemia associated with t(7;8)(q34;p11.2) and FGFR1 rearrangement. This report was approved by the Institutional Review Board of Kyung Hee Medical Center, Seoul, Korea (KHUH 2019-09-057) and granted a waiver of informed consent as this was a retrospective study using residual samples.
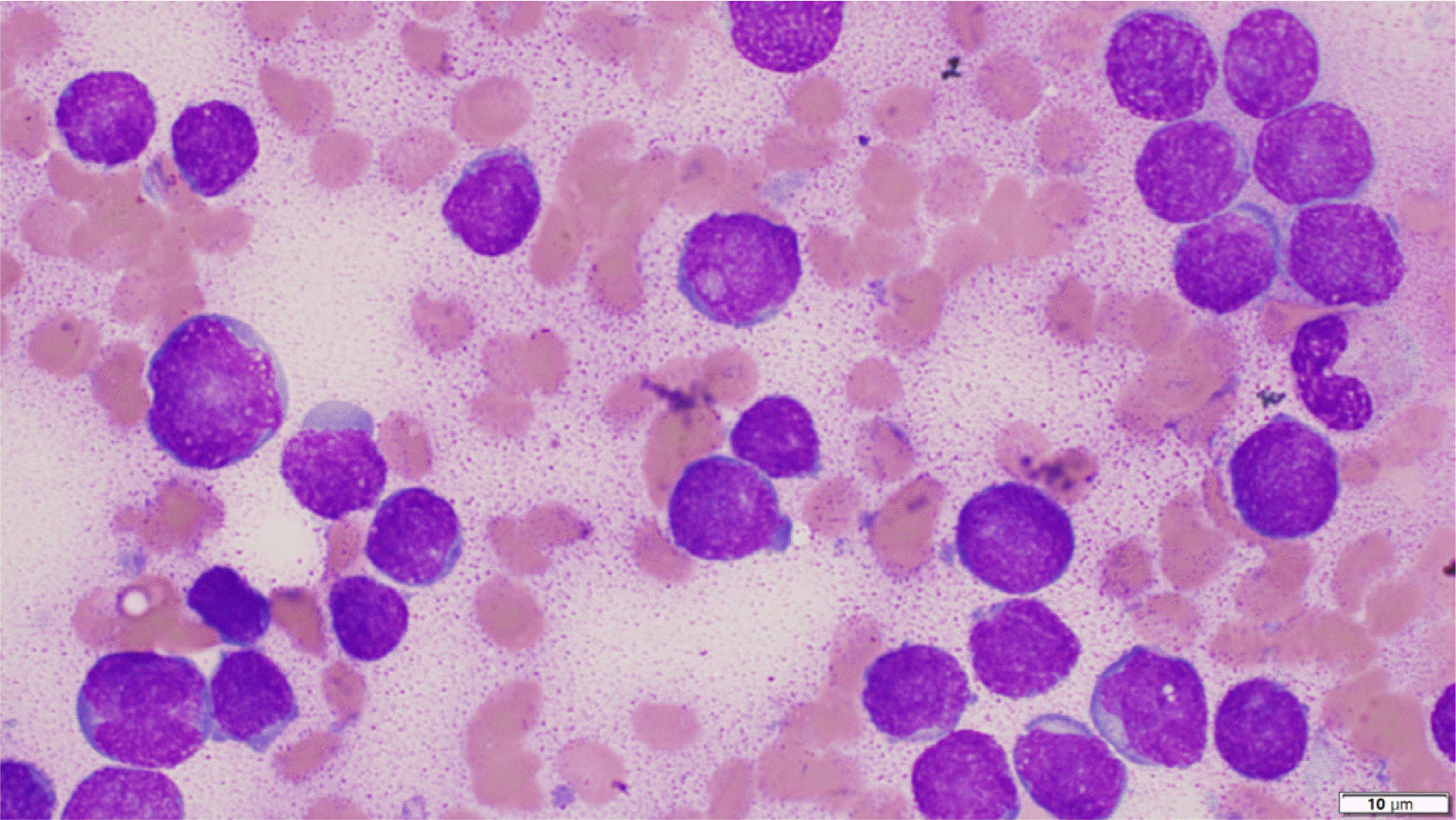
A 24-month-old male patient was admitted to our hospital in January 2019 with fever up to 39°C, which started eight days before the visit, and petechiae on the upper chest. A complete blood count revealed white blood cell (WBC) count 7.32×109/L, Hb 91 g/L, and platelets 36×109/L. Initial WBC differential counts were 10% segmented neutrophils, 67% lymphocytes, 9% monocytes, 6% atypical lymphocytes, 1% metamyelocytes, 1% myelocytes, and 6% blasts. A bone marrow (BM) aspirate showed a hypercellular marrow with an increased number of blasts (86.2% of all nucleated cells) (Fig. 1).
In a biopsy section of the BM, the estimated cellularity was 80%–90%. A BM chromosome study revealed 46,XY,t(7;8)(q34;p11.2),i(9)(q10) in 16 out of 20 metaphase cells analyzed (Fig. 2A). The patient was suspected of having FGFR1 rearrangement. Therefore, FISH analysis (MetaSystems, Altlussheim, Germany) using an FGFR1 probe was performed, and break-apart signals were observed in 106 out of 300 interphase cells (35.3%) (Fig. 2B). Genetic rearrangement was not detected by multiplex reverse transcription PCR using cDNA, mastermix and split-out PCR primer mix from the HemaVision kit (DNA Diagnostic A/S, Risskov, Denmark) on the SimpliAmp thermal cycler (Thermo Fisher Scientific, Waltham, MA, USA), following the manufacturer’s instructions.
Flow cytometric analysis (Navios EX Flow Cytometer and Kaluza Analysis Software, Beckman Coulter, Miami, FL, USA) of BM specimens performed per the manufacturer’s instructions revealed that blasts expressed CD34, CD10, CD19, CD33, CD20, CD38, cCD22, cCD79a, and TdT. Finally, the patient was diagnosed as having B-lymphoblastic leukemia with FGFR1 rearrangement. The patient was transferred to another hospital immediately after diagnosis and no further clinical and laboratory results were obtained.
FGFR1 is associated with clonal eosinophilic disorders and can undergo various gene rearrangements with at least 44 partner genes [2]. Gene rearrangement associated with FGFR1 has been termed “8p11 myeloproliferative syndrome (EMS)” in the past because of the chromosomal location of FGFR1, (8p11-12) [3]. Currently, it is classified as an independent myeloid/lymphoid neoplasm group [1]. The clinical features of myeloid/lymphoid neoplasm with FGFR1 rearrangement can take many forms, including chronic eosinophilic leukemia, AML, T-cell lymphoblastic lymphoma, and occasionally, precursor B-lymphoblastic leukemia/lymphoma [1, 4]. Patients in the chronic phase of myeloid/lymphoid neoplasm with FGFR1 rearrangement usually show eosinophilia and neutrophilia and sometimes, monocytosis [4]. The most common FGFR1 rearrangement is related to t(8;13)(p11.2;q12.1), where the ZMYM2-FGFR1 fusion gene can be detected at the molecular level [5]. In addition, FGFR1 rearrangement associated with approximately 10 different partner genes is listed in the 2016 WHO classification [1].
To the best of our knowledge, t(7;8)(q34;p11.2) is a very rare FGFR1 rearrangement and has been reported in only one AML case in a 49-year-old Italian female patient [6]. Furthermore, our case is the first report of t(7;8)(q34;p11.2) in lymphoid malignancies. In the Italian AML patient, a TRIM24-FGFR1 fusion gene was identified at the molecular level [6]. However, our patient was transferred to another hospital and was lost to follow-up. TRIM24 (also known as TIF1α) encodes a nuclear protein that contributes to polymerization and ubiquitylation and thus, regulates developmental and physiologic processes [7, 8]. TRIM24 proteins have various functions in transcription, cell differentiation, mitosis, and DNA repair, and functional abnormalities can lead to tumorigenesis [7, 9]. Malignant disorders associated with TRIM24 abnormalities include thyroid, breast, lung, liver, colorectal, and prostate cancers [7-9]. In most cancers, overexpressed TRIM24 acts as an oncogene associated with cancer progression; however, in hepatocellular carcinoma, it functions as a tumor suppressor [8, 9]. TRIM24 abnormalities in hematologic malignancies have not been sufficiently studied and further studies will be necessary.
Until recently, the prognosis of hematologic malignancy associated with FGFR1 rearrangement was very poor, and stem cell transplantation was the only treatment option [4, 5]. However, tyrosine kinase inhibitors have enabled molecular targeted therapy in BCR-ABL1-positive chronic myeloid leukemia, which was incurable in the past [10]. Similarly, we believe that consistent clinical reporting of rare gene rearrangements will facilitate the development of personalized treatments in future.
Go to : 
Notes
AUTHOR CONTRIBUTIONS
Cho H and Han Y coordinated the drafting of the manuscript and wrote the manuscript. Lee JJ collected the data. Park KS, Kim YJ, and Cho SY interpreted the data. Yoon HS managed the patient and provided the clinical information. Lee WI reviewed and commented on the draft. Park TS designed and supervised the study. All authors have accepted their responsibility for the entire content of this manuscript and approved submission.
Go to : 
REFERENCES
1. Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. 2016; The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 127:2391–405. DOI: 10.1182/blood-2016-03-643544. PMID: 27069254.

2. Huret JL. 2009; FGFR1 (fibroblast growth factor receptor 1). Atlas of Genetics and Cytogenetics in Oncology and Haematology. 13:821–40. DOI: 10.1161/HYPERTENSIONAHA.120.15587,. PMID: 33131311.

3. Macdonald D, Reiter A, Cross NC. 2002; The 8p11 myeloproliferative syndrome: a distinct clinical entity caused by constitutive activation of FGFR1. Acta Haematol. 107:101–7. DOI: 10.1159/000046639. PMID: 11919391.

4. Bain BJ, Horny HP, Arber DA, Tefferi A, Hasserjian RP. Swerdlow SH, Campo E, editors. 2017. Myeloid/lymphoid neoplasms with eosinophilia and rearrangement of PDGFRA, PDGFRB or FGFR1, or with PCM1-JAK2. WHO classification of tumours of haematopoietic and lymphoid tissues. 4th ed. International Agency for Research on Cancer;Lyon: p. 72–9.
5. Jackson CC, Medeiros LJ, Miranda RN. 2010; 8p11 myeloproliferative syndrome: a review. Hum Pathol. 41:461–76. DOI: 10.1016/j.humpath.2009.11.003. PMID: 20226962.

6. Belloni E, Trubia M, Gasparini P, Micucci C, Tapinassi C, Confalonieri S, et al. 2005; 8p11 myeloproliferative syndrome with a novel t(7;8) translocation leading to fusion of the FGFR1 and TIF1 genes. Genes, Chromosomes and Cancer. 42:320–5. DOI: 10.1002/gcc.20144. PMID: 15609342.
7. Hu Q, Wang C, Xiang Q, Wang R, Zhang C, Zhang M, et al. 2020; Discovery and optimization of novel N-benzyl-3,6-dimethylbenzo[d]isoxazol-5-amine derivatives as potent and selective TRIM24 bromodomain inhibitors with potential anti-cancer activities. Bioorg Chem. 94:103424. DOI: 10.1016/j.bioorg.2019.103424. PMID: 31776034.

8. Huret JL. 2007; TRIM24 (tripartite motif-containing 24). Atlas of Genetics and Cytogenetics in Oncology and Haematology. 11:111–2.
9. McAvera RM, Crawford LJ. 2020; TIF1 proteins in genome stability and cancer. Cancers (Basel). 12:2094. DOI: 10.3390/cancers12082094. PMID: 32731534. PMCID: PMC7463590.

10. Hughes T, Deininger M, Hochhaus A, Branford S, Radich J, Kaeda J, et al. 2006; Monitoring CML patients responding to treatment with tyrosine kinase inhibitors: review and recommendations for harmonizing current methodology for detecting bcr-abl transcripts and kinase domain mutations and for expressing results. Blood. 108:28–37. DOI: 10.1182/blood-2006-01-0092. PMID: 16522812. PMCID: PMC1895821.

Go to : 




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download