INTRODUCTION
MATERIALS AND METHODS
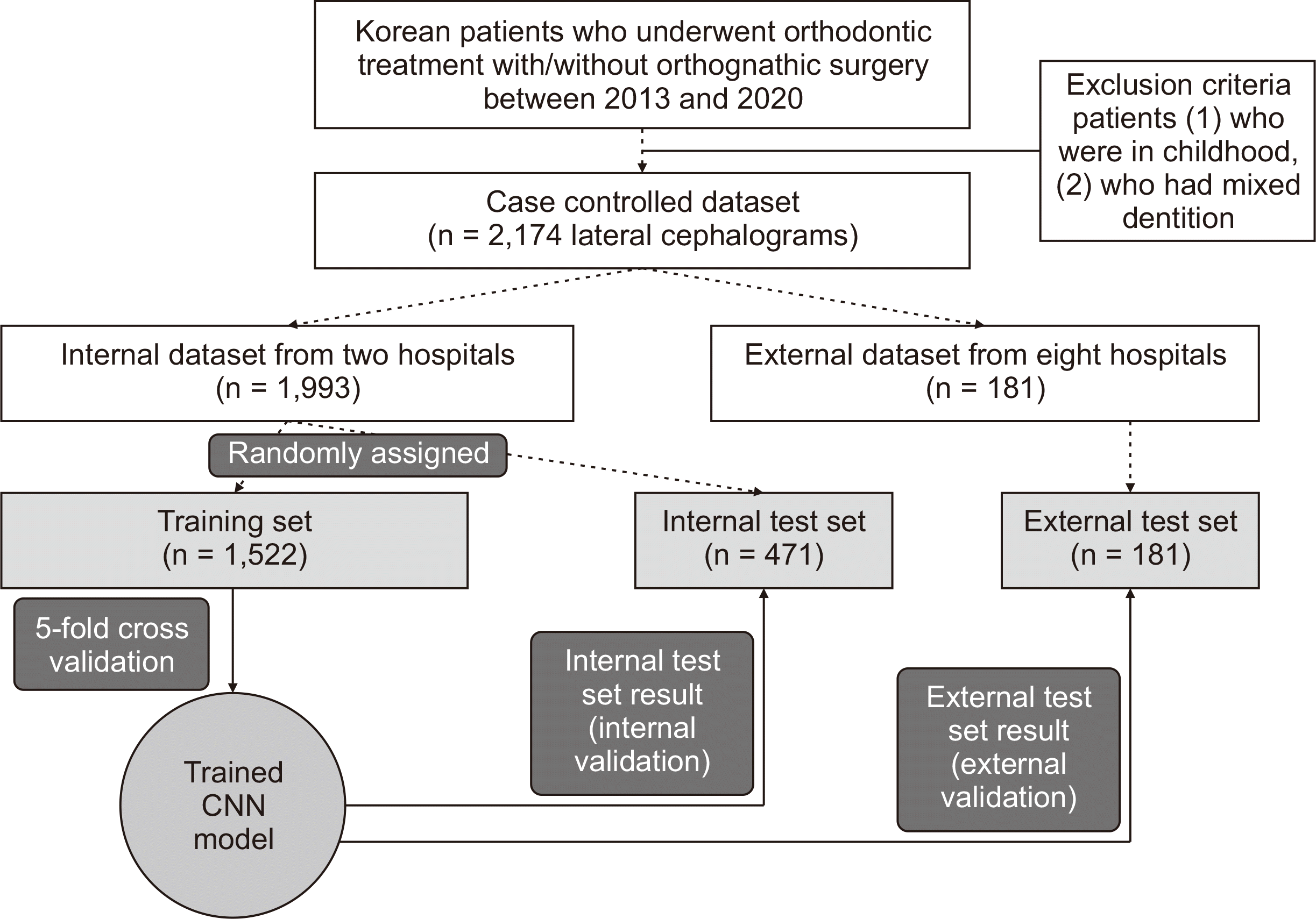
Description of the dataset
Table 1
SNUDH, Seoul National University Dental Hospital; KADH, Kooalldam Dental Hospital; AJUDH, Ajou University Dental Hospital; AMC, Asan Medical Center; CNUDH, Chonnam National University Dental Hospital; CSUDH, Chosun University Dental Hospital; EUMC, Ewha University Medical Center; KHUDH, Kyung Hee University Dental Hospital; KNUDH, Kyungpook National University Dental Hospital; WKUDH, Wonkwang University Dental Hospital; CR, computed radiography; CCD, charge-coupled device.
Setting a gold standard for the diagnosis of APSDs, VSDs, and VDDs
Table 2
| Sex | APSDs | VSDs | VDDs | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ANB | FMA | FHR | Overbite | ||||||||
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | ||||
| Female | 2.4 | 1.8 | 24.2 | 4.6 | 65 | 9 | 1.5 | 1.5 | |||
| Male | 1.78 | 2.02 | 26.78 | 1.79 | 66.37 | 5.07 | |||||
Table 3
APSDs, anteroposterior skeletal discrepancies; VSDs, vertical skeletal discrepancies; VDDs, vertical dental discrepancies; SNUDH, Seoul National University Dental Hospital; KADH, Kooalldam Dental Hospital; AJUDH, Ajou University Dental Hospital; AMC, Asan Medical Center; EUMC, Ewha University Medical Center; CNUDH, Chonnam National University Dental Hospital; CSUDH, Chosun University Dental Hospital; KHUDH, Kyunghee University Dental Hospital; KNUDH, Kyungpook National University Dental Hospital; WKUDH, Wonkwang University Dental Hospital.
Preprocessing of the data
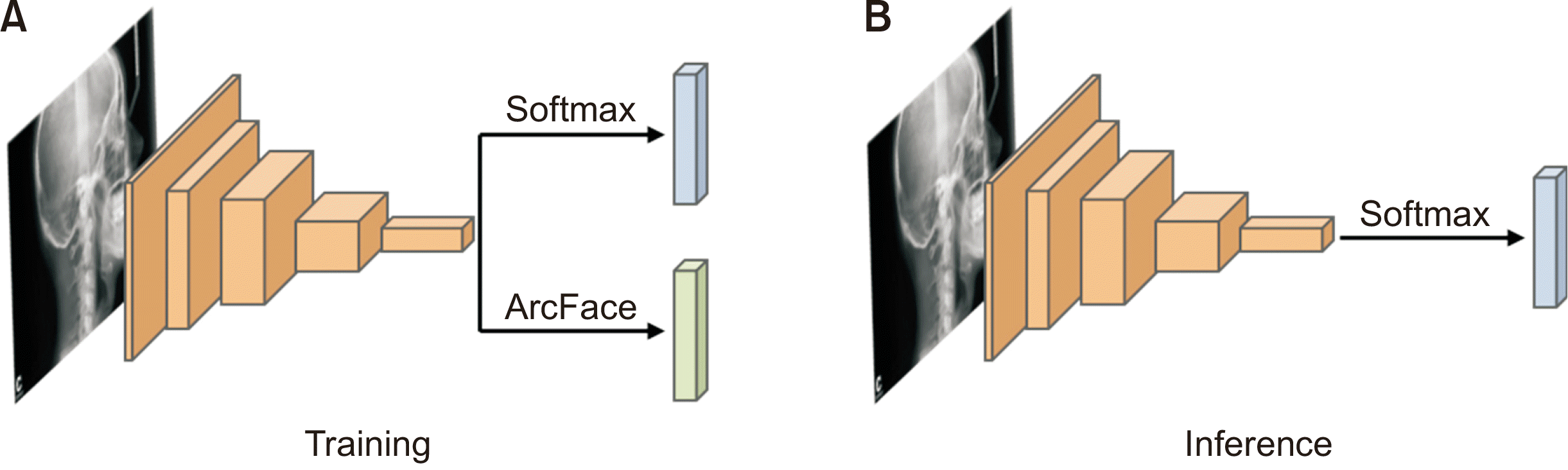
Model architecture (Figure 2)
Model training (Figures 1 and 2)
Model testing
Analysis method
t-stochastic neighbor embedding (t-SNE)
Gradient-weighted class activation mapping (Grad-CAM)25
RESULTS
Metrology distribution of the APSDs, VSDs, and VDDs per dataset (Figure 3)
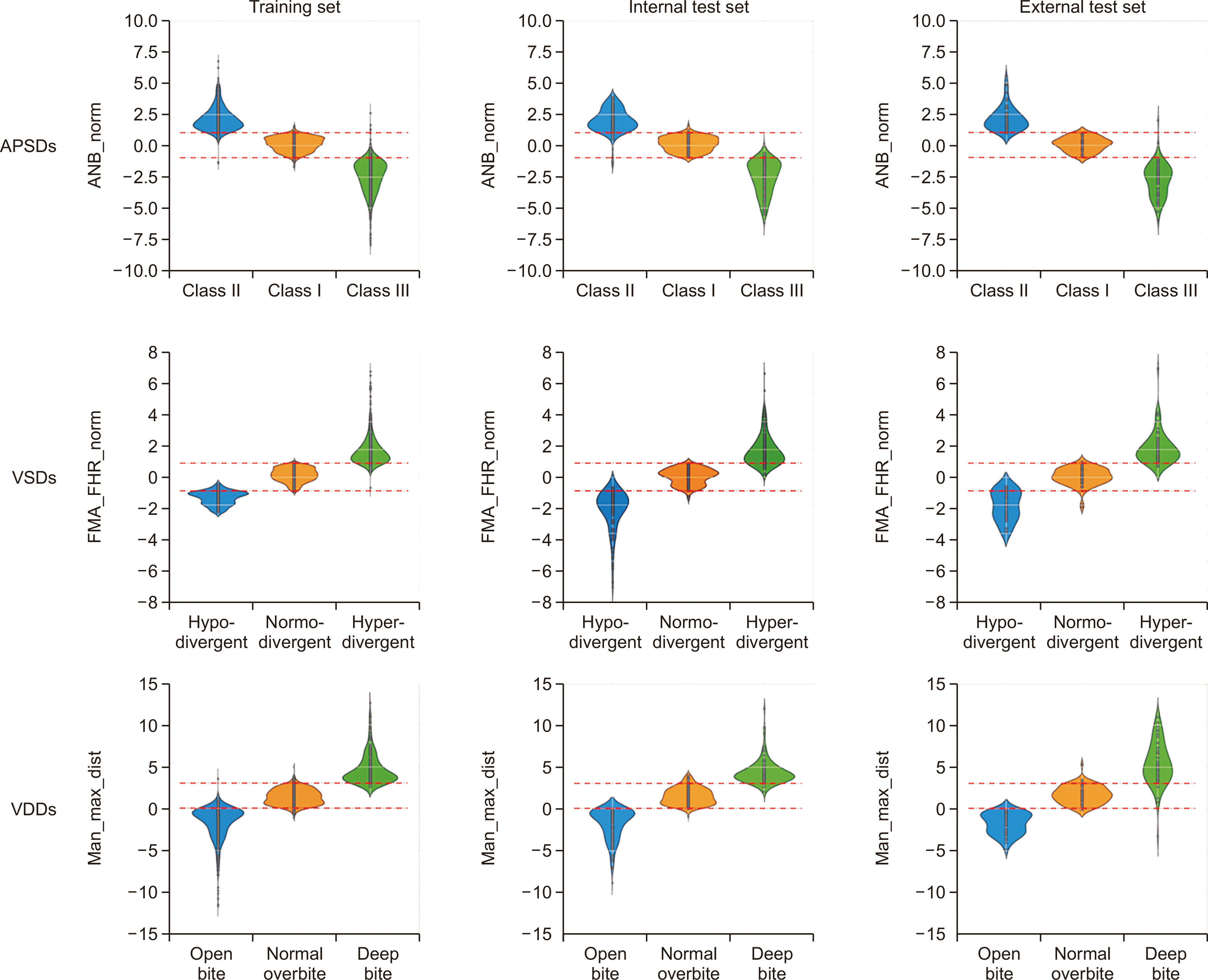 | Figure 3Metrology distribution of the anteroposterior skeletal discrepancies (APSDs: Class I, Class II, and Class III), vertical skeletal discrepancies (VSDs: normodivergent pattern, hyperdivergent pattern, and hypodivergent pattern), and vertical dental discrepancies (VDDs: normal overbite, open bite, and deep bite) per dataset. Red lines in APSDs and VSDs indicate one standard deviation of the normal classification. Red lines in VDDs indicate the boundary values, which were 0 mm and 3 mm.
ANB, angle among A point, nasion, and B point; FMA, Frankfort mandibular plane angle; FHR, Jarabak’s posterior/anterior facial height ratio; norm, normalized; Man, mandible 1 crown; Max, maxilla 1 crown; dist, distance.
|
Accuracy and AUC of the internal test set in binary ROC analysis (Table 4 and Figure 4)
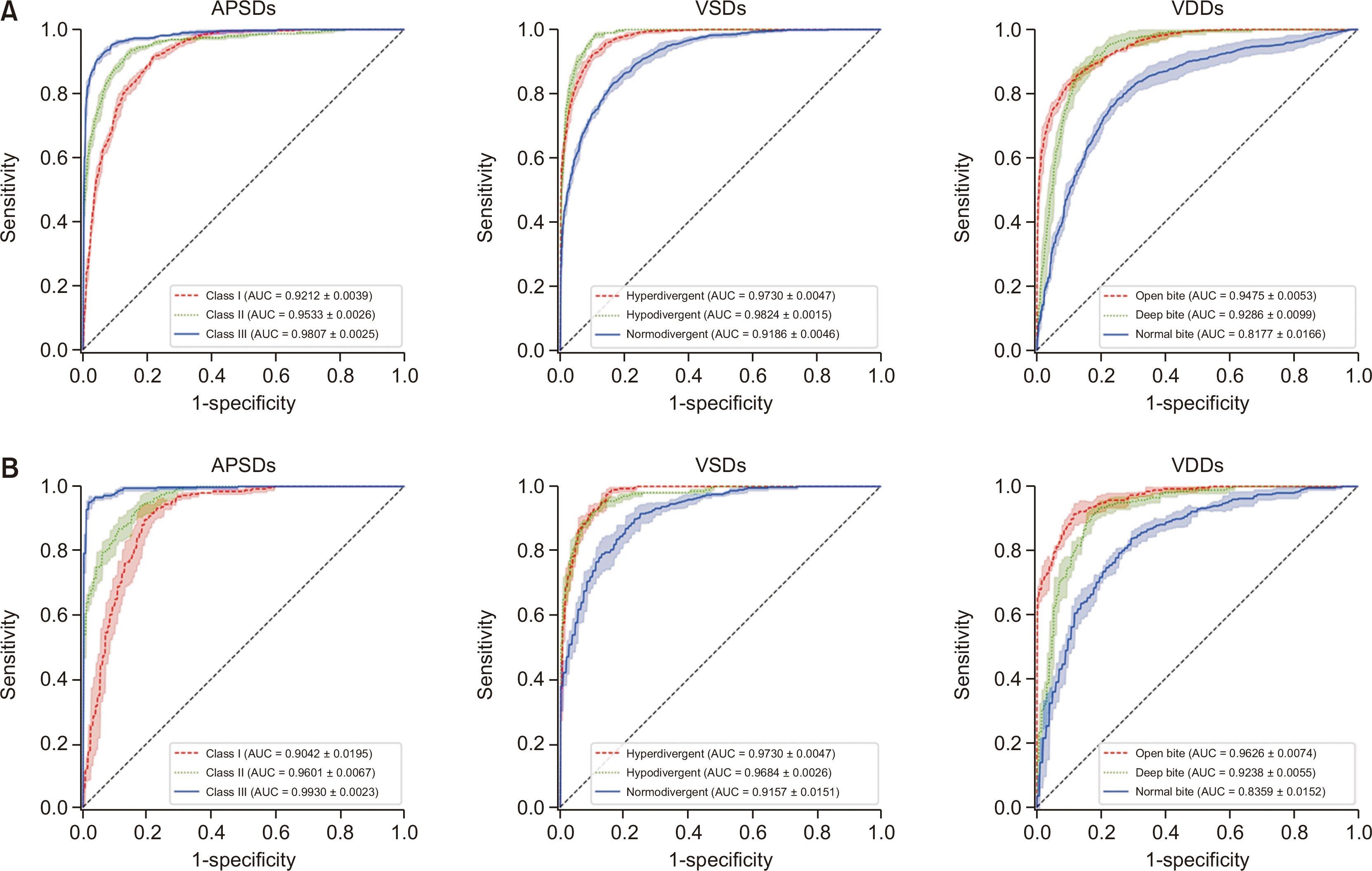 | Figure 4The results of the binary receiver operating characteristic curve analysis (A) in the internal test set from two hospitals and (B) in the external test set from other 8 hospitals for diagnosis of anteroposterior skeletal discrepancies (APSDs), vertical skeletal discrepancies (VSDs), and vertical dental discrepancies (VDDs).
AUC, area under the curve.
|
Table 4
Accuracy and AUC of the external test set in binary ROC analysis (Table 4 and Figure 4)
Comparison of AUC values between internal and external test sets in binary ROC analysis (Table 4)
Comparison of AUC values between internal and external test sets in multiple ROC analysis (Table 5)
Table 5
APSDs, anteroposterior skeletal discrepancies; VSDs, vertical skeletal discrepancies; VDDs, vertical dental discrepancies; ROC, receiver operating characteristic; AUC, area under the curve; SD, standard deviation; Hyper, hyperdivergent; Hypo, hypodivergent; Normo, normodivergent; Open, open bite; Deep, deep bite; Normal, normal overbite.
t-SNE of APSDs, VSDs, and VDDs per dataset (Figure 5)
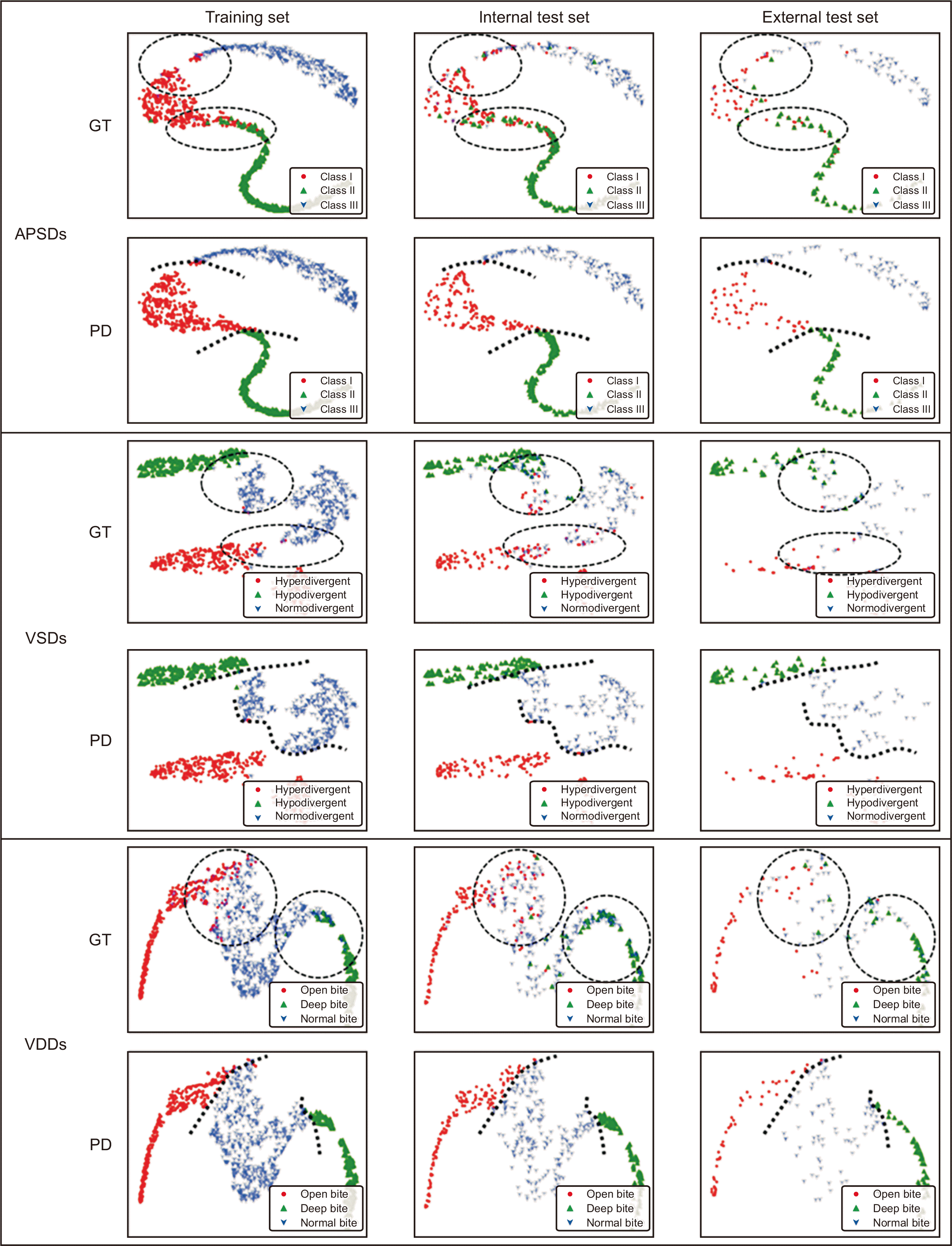 | Figure 5The results of t-stochastic neighbor embedding in anteroposterior skeletal discrepancies (APSDs), vertical skeletal discrepancies (VSDs), and vertical dental discrepancies (VDDs) per dataset. The labels of ground truth (GT) and prediction (PD) were set to check their distribution. Dotted circles indicate areas with irregular mixing. Dotted lines indicate cutoff lines. |
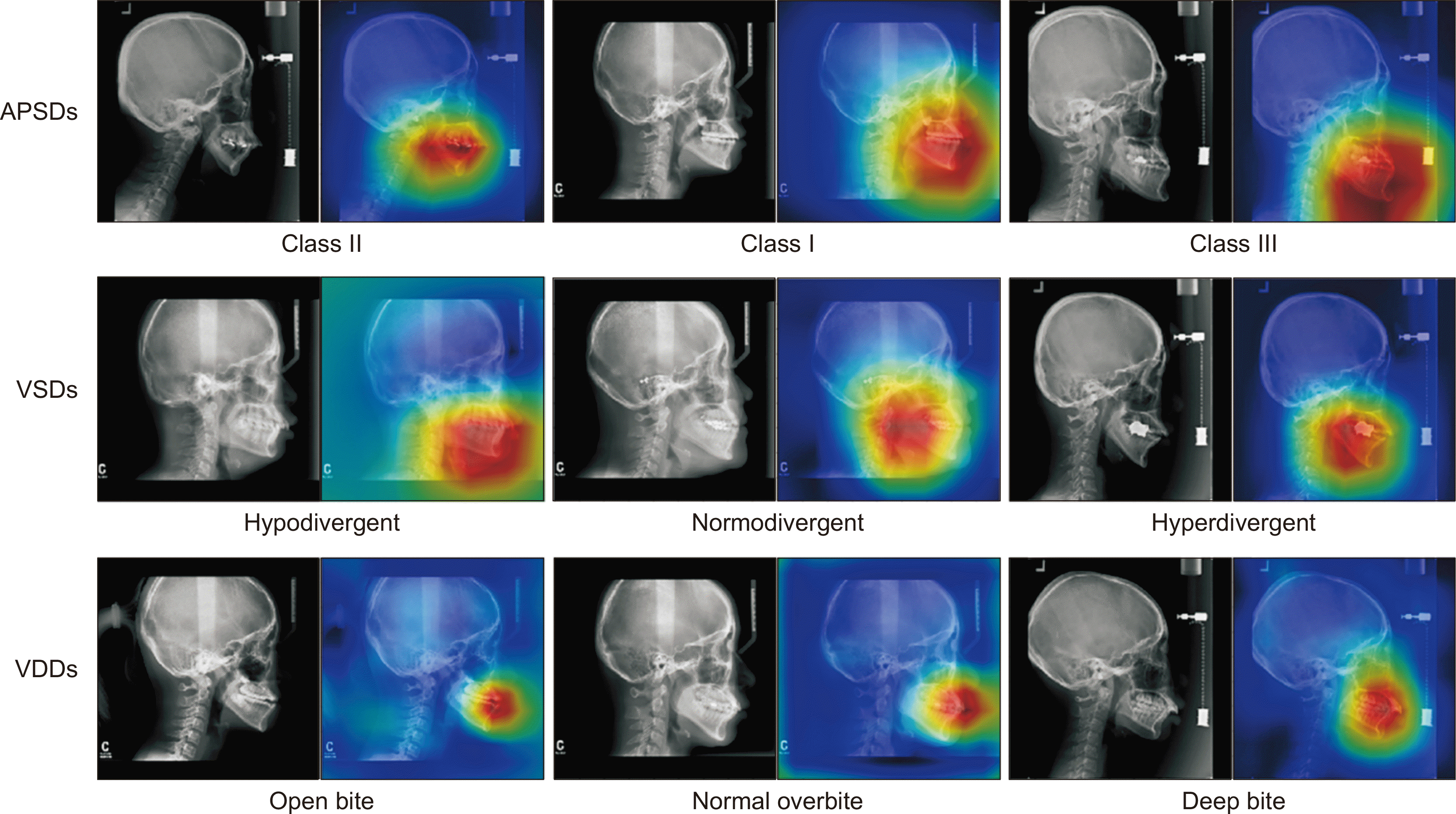
Grad-CAM for each diagnosis (Figure 6)
DISCUSSION
Clinical meaning of the comparison results between internal and external test sets in binary and multiple ROC analysis
Comparison of accuracy with a previous study using binary ROC analysis results
Table 6
| Models | APSDs | VSDs | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Accuracy | AUC | Sensitivity | Specificity | Accuracy | AUC | ||||||||||||||||
|
Yu et al’s study8 |
This study | Yu et al’s study8 | This study | Yu et al’s study8 | This study | Yu et al’s study8 | This study | Yu et al’s study8 | This study | Yu et al’s study8 | This study | Yu et al’s study8 | This study | Yu et al’s study8 |
This study |
||||||||
|
Model I (no exclusion of data set) |
0.8575 | 0.8414 | 0.9288 | 0.9206 | 0.9050 | 0.8944 | 0.938 | 0.9517 | 0.8427 | 0.8461 | 0.9213 | 0.9146 | 0.8951 | 0.8910 | 0.937 | 0.9580 | |||||||
|
Model II (exclusion of data set within interval of 0.2 SD) |
0.9079 | NA | 0.9539 | NA | 0.9386 | NA | 0.970 | NA | 0.9222 | NA | 0.9611 | NA | 0.9481 | NA | 0.985 | NA | |||||||
|
Model III (exclusion of data set within interval of 0.3 SD) |
0.9355 | NA | 0.9677 | NA | 0.9570 | NA | 0.978 | NA | 0.9459 | NA | 0.9729 | NA | 0.9640 | NA | 0.984 | NA | |||||||
Difference in the AUC values of in Class II and Class III groups in APSDs and hyperdivergent and hypodivergent groups in VSDs in binary and multiple ROC analysis
Lower AUC values in VDDs compared to APSDs and VSDs in binary ROC analysis
Current status of CNN-based orthodontic diagnosis
Table 7
| Author (year) | Samples | Model and its application | Data set | Results |
|---|---|---|---|---|
|
Arık et al. (2017)1 |
||||
|
Park et al. (2019)9 |
|
|||
|
Nishimoto et al. (2019)3 |
||||
|
Hwang et al. (2020)10 |
|
|||
|
Kunz et al. (2020)11 |
||||
|
Yu et al. (2020)8 |
<Model I> Sagittal Vertical |
|
||
|
Kim et al. (2020)2 |
Evaluation group 1: Evaluation group 2: Evaluation group 3: |
|||
|
This study (2020) |
|
CNN, convolutional neural network; YOLO, “you only look once” real-time object detection; SSD, single shot detector; ISBI, International Symposium on Biomedical Imaging; AI, artificial intelligence; CI, confidence interval; ROC, receiver operating characteristic; t-SNE, t-stochastic neighbor embedding; Grad-CAM, gradient-weighted class activation mapping.
Limitations of this study and suggestions for future studies
CONCLUSION
The accuracy of our model was well-validated with internal test sets from two hospitals as well as external test sets from eight other hospitals without issues regarding the continuity of the data sets or exaggerated accuracy.
Our model shows the possible usefulness of a one-step automated orthodontic diagnosis tool for classifying skeletal and dental discrepancies with input of lateral cephalograms only in an end-to-end manner. However, it still needs technical improvement in terms of classifying VDDs.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download