Introduction
Materials and Methods
Maintenance of human pluripotent stem cells (hPSCs)
Differentiation of hESCs into dopaminergic neurons, osteoblasts, and hepatocytes
Total RNA extraction and qRT-PCR
Table 1.
Statistics
Results
Conservative expression of imprinted genes in undifferentiated hPSC lines
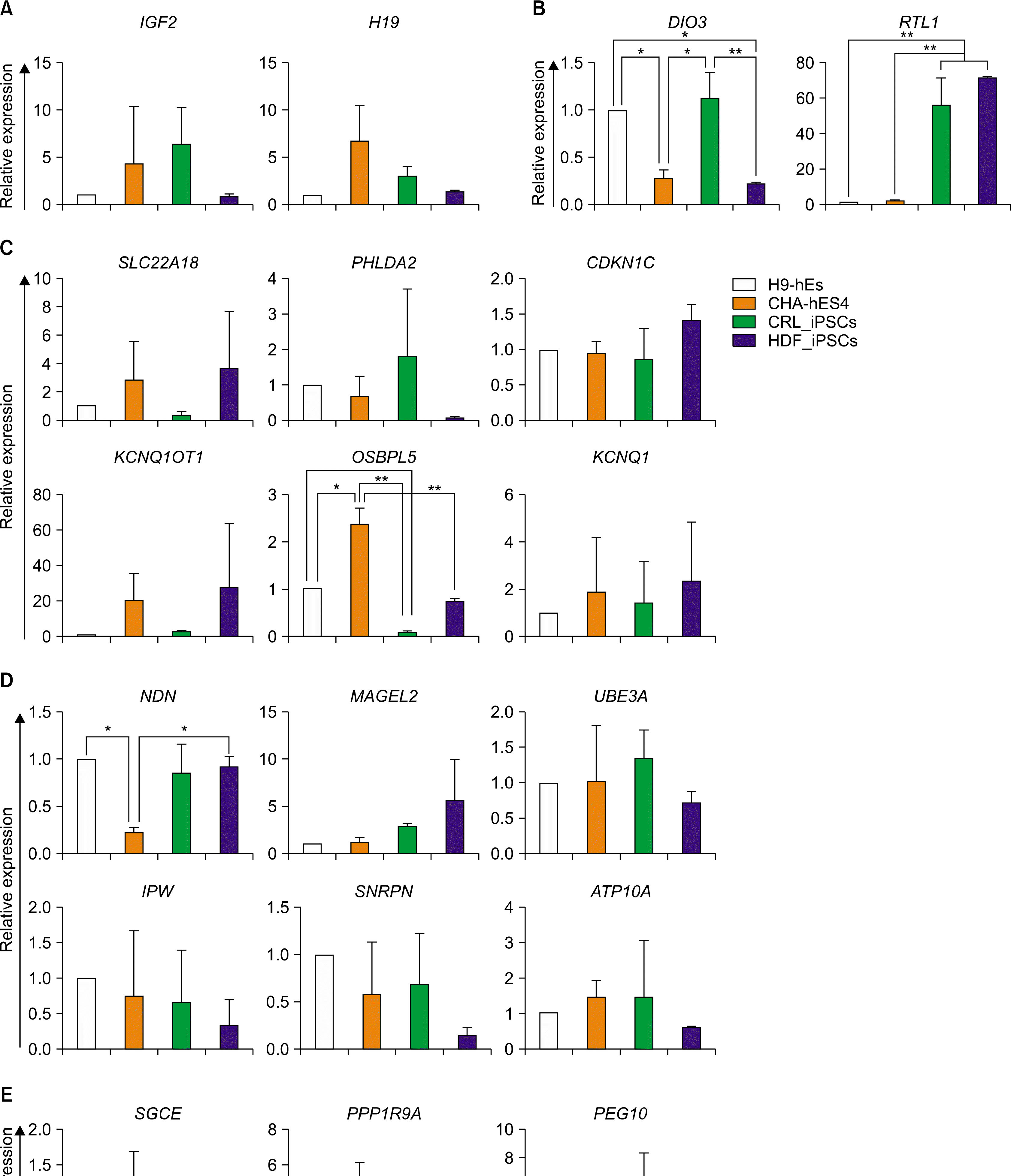 | Fig. 1.Transcriptional expression of imprinted genes between four hPSC lines. Transcriptional levels of the imprinted genes that are included in IGF2-H19 (A), DIO3-DLK1 (B), KCNQ1 (C), PWS/AS (D), and PEG10 (E) domains were examined in undifferentiated hPSCs by qRT-PCR. The error bars indicate±SD for three independent experiments (n=3, *p<0.05, **p <0.01). Abbreviations: CRL_iPSCs; hiPSCs derived from CRL foreskin fibroblasts, HDF_iPSCs; hiPSCs derived from human dermal fibro-blasts. |
Quantitative expression of the imprinted genes in hESCs during in vitro differentiation
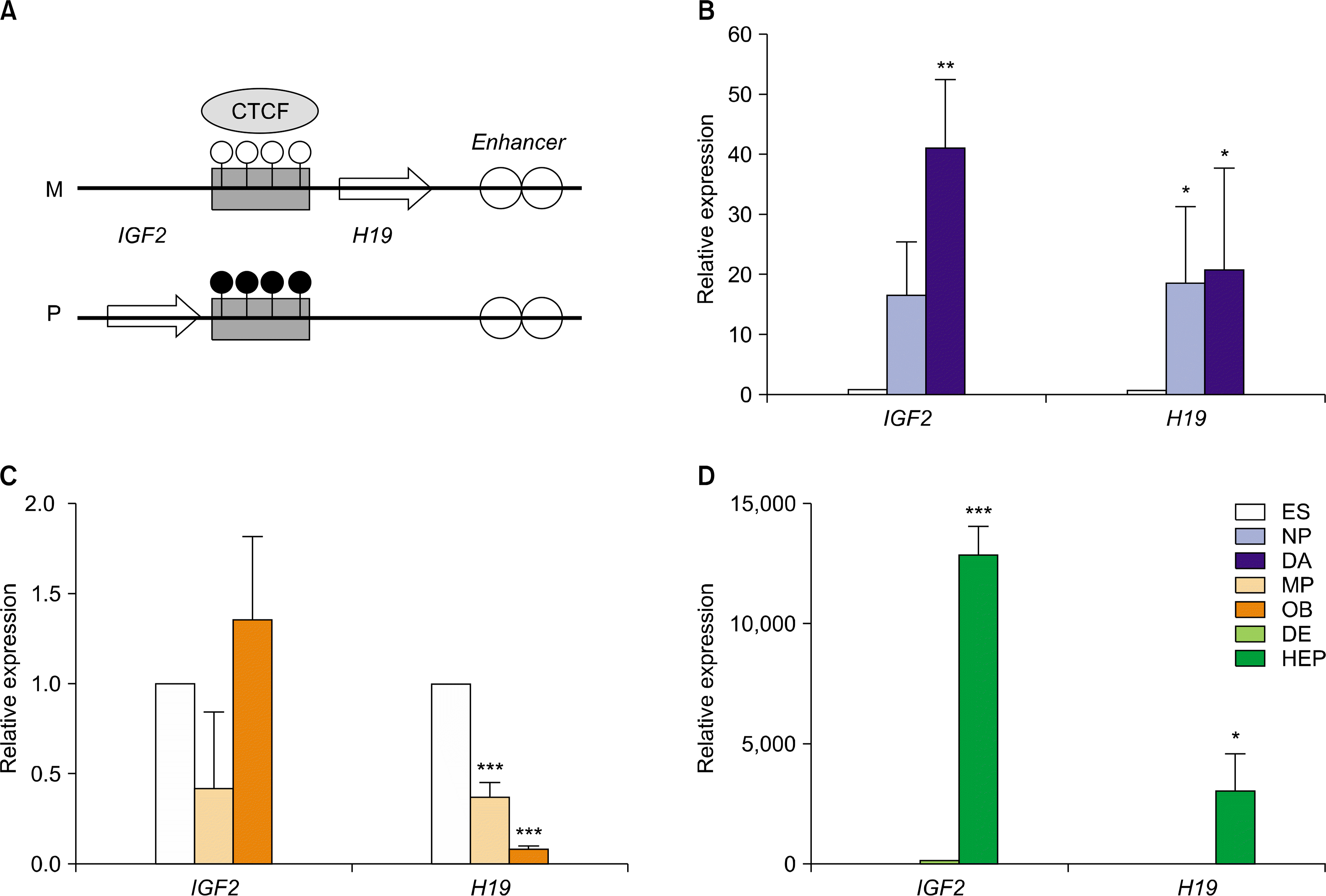 | Fig. 2.Relative expression levels of imprinted genes in IGF2/H19 domain in hESCs during in vitro differentiation. (A) A schematic diagram of IGF2/H19 domain (Ch 11p15). Relative expression level of IGF and H19 imprinted genes in hESCs during in vitro differentiation to DA neurons (B), osteoblasts (C), and hepatocytes (D). The error bars indicate±SD for three independent experiments (n=3, *p<0.05, **p<0.01, ***p<0.001). Abbreviations, ES: H9-hESCs; NP: neuronal progenitors; DA: dopaminergic neurons; MP: mesenchymal progenitors; OB: osteoblasts; DE: definitive endoderm; HEP: hepatocytes. |
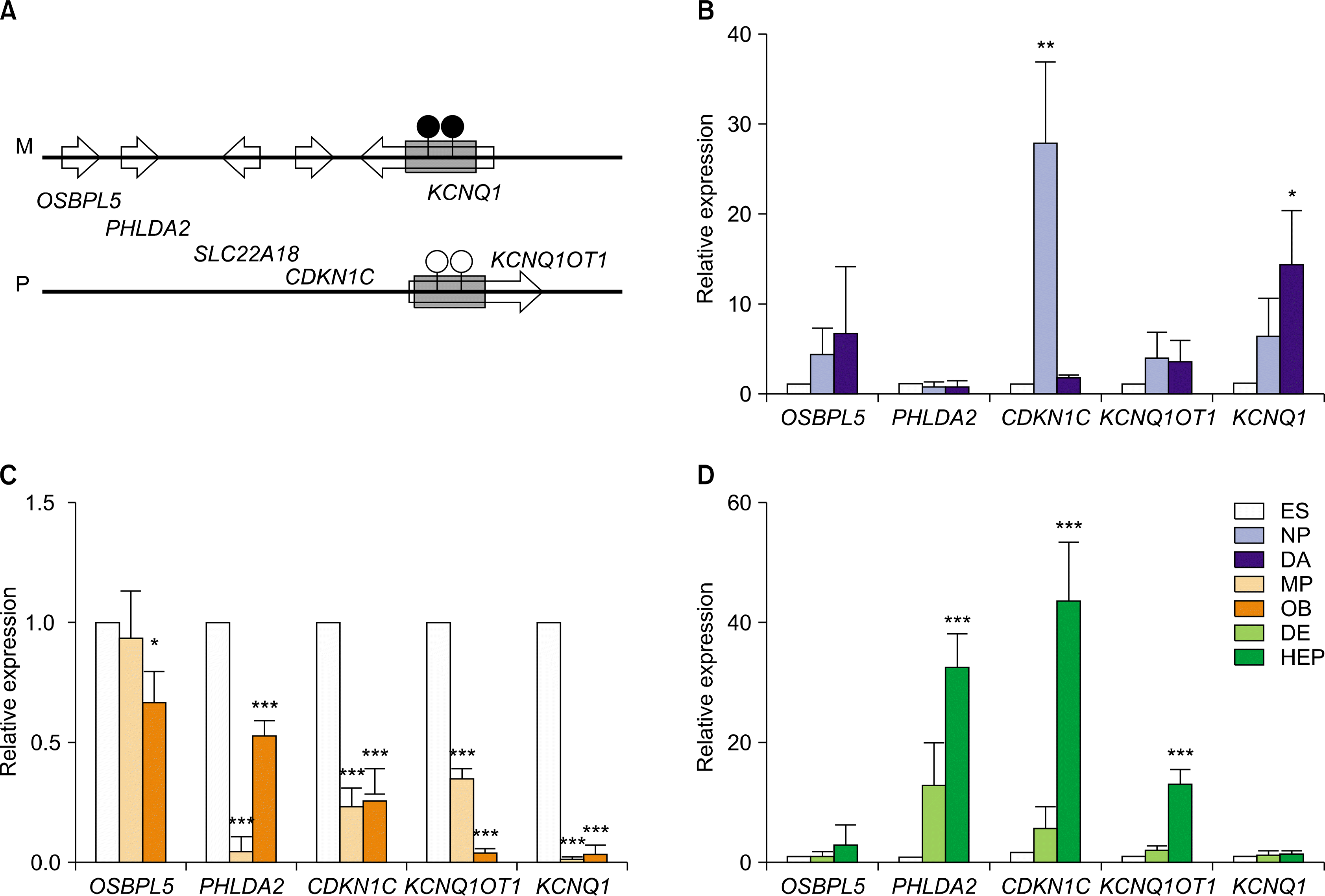 | Fig. 3.Relative expression levels of imprinted genes in KCNQ1 domain in hESCs during in vitro differentiation. (A) A schematic diagram of KCNQ1 domain (Ch 11p15). This KCNQ1 domain contains several imprinted genes, including OSBPL5, PHLDA2, CDKN1C, KCNQ1, and KCNQ1OT1. Relative expression level of several imprinted genes in hESCs during in vitro differentiation to DA neurons (B), osteoblasts (C), and hepatocytes (D). The error bars indicate±SD for three independent experiments (n=3, *p<0.05, **p<0.01, ***p<0.001). |
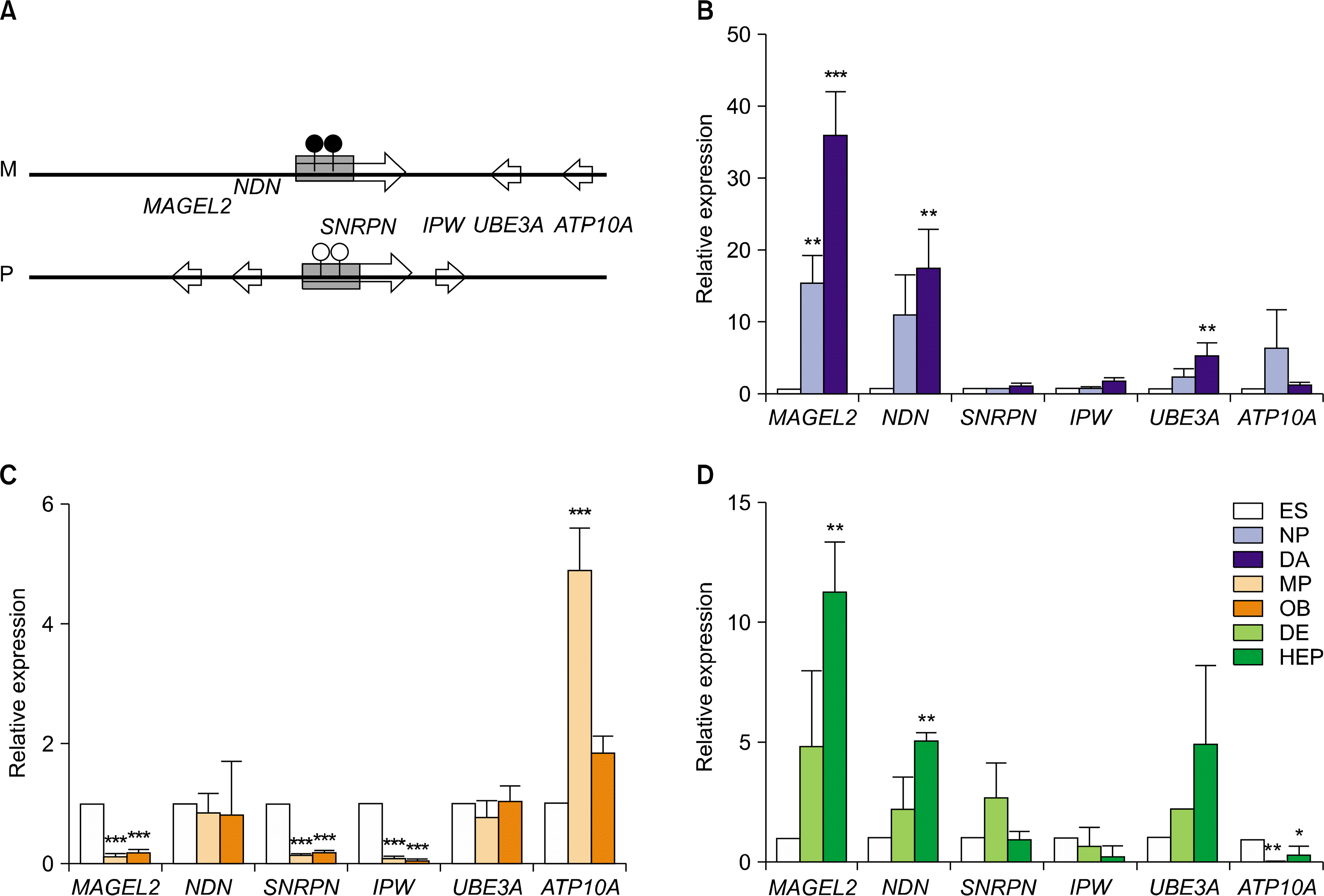 | Fig. 4.Relative expression levels of imprinted genes in PWS/AS domain in hESCs during in vitro differentiation. (A) A schematic diagram of PWS/AS domain (Ch. 15q11-q13). This domain contains several imprinted genes, including MAGEL2, NDN, SNRPN, IPW, UBE3A, and ATP10A. Relative expression level of several imprinted genes in hESCs during in vitro differentiation to DA neurons (B), osteoblasts (C), and hepatocytes (D). The error bars indicate±SD for three independent experiments (n=3, *p<0.05, **p<0.01, ***p<0.001). |
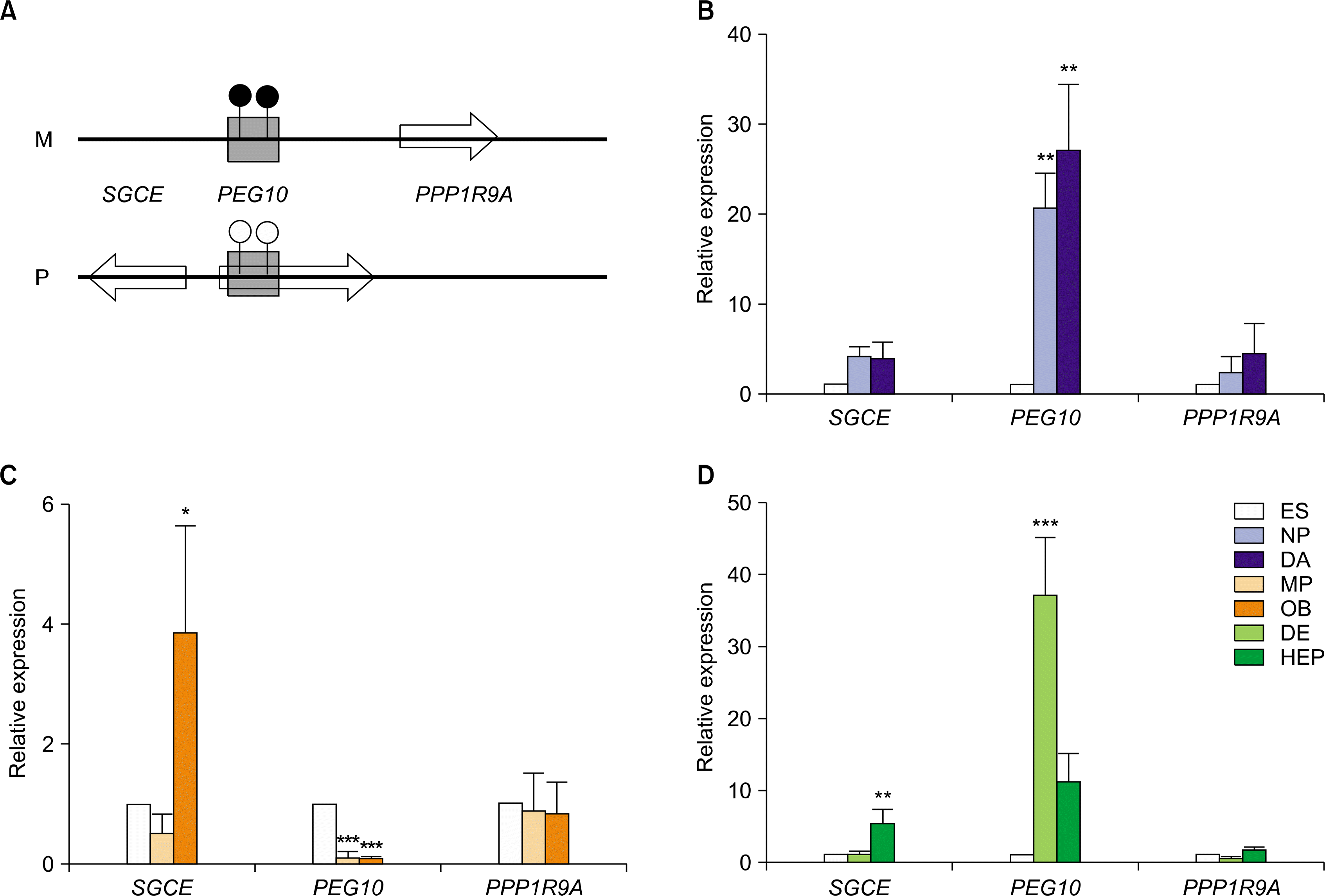 | Fig. 5.Relative expression levels of imprinted genes in PEG10 domain in hESCs during in vitro differentiation. (A) A schematic diagram of PEG10 domain (Ch. 7q21.3). This domain contains three imprinted genes such as SGCE, PEG10 and PPP1R9A. Relative expression level of these imprinted genes in hESCs during in vitro differentiation to DA neurons (B), osteoblasts (C), and hepatocytes (D). The error bars indicate±SD for three independent experiments (n=3, *p<0.05, **p<0.01, ***p<0.001). |
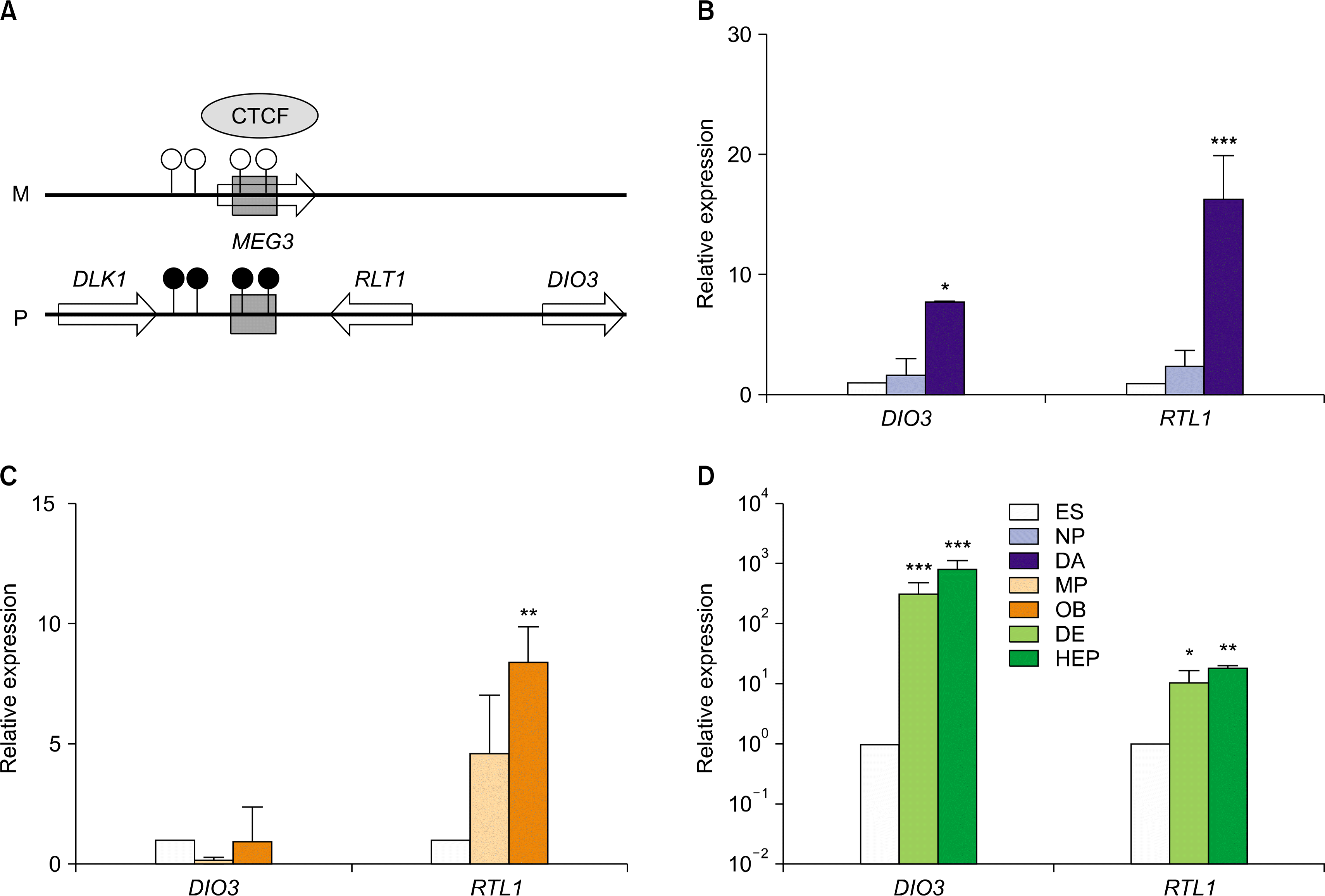 | Fig. 6.Relative expression levels of imprinted genes in DIO3-DLK1 domain in hESCs during in vitro differentiation. (A) A schematic diagram of DIO3-DLK1 domain (Ch. 14q32.2). This domain contains four imprinted genes such as DLK1, MEG3, DIO3 and RTL1. Relative expression level of DIO3 and RTL1 imprinted genes in hESCs during in vitro differentiation to DA neurons (B), osteoblasts (C), and hepatocytes (D). The error bars indicate±SD for three independent experiments (n=3, *p<0.05, **p<0.01, ***p<0.001). |




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download