Introduction
Materials and Methods
Drugs and chemicals
Animals
Group 1 (Control Group)
Group 2 (LPS Group)
Group 3 (LPS and TQ Group)
Group 4 (LPS and PNU-120596 Group)
Subgroup 4a
Subgroup 4b
Immunohistochemical Study
Anti-phospho-cyclic adenosine phosphate (cAMP) response element binding protein (P-CREB) (11) immunostaining, a beta transcription factor that activates target genes through cAMP response elements, CREB plays a key role in neuronal survival, precursor proliferation and neuronal differentiation. Primary antibody (Ab) rabbit polyclonal supplier (Merck Milipore, Germany) catalog number (06-519) used as 0.1 ml diluted stored at 2~8°C.
CD44 Ab to detect the endogenous MSCs (12). 7 ml prediluted rabbit monoclonal 1ry Ab (IW-PA1021) (IHC World, Ellicott City, USA) stored at 2~8°C.
Morphometric Study
Statistical analysis
Results
Histological results
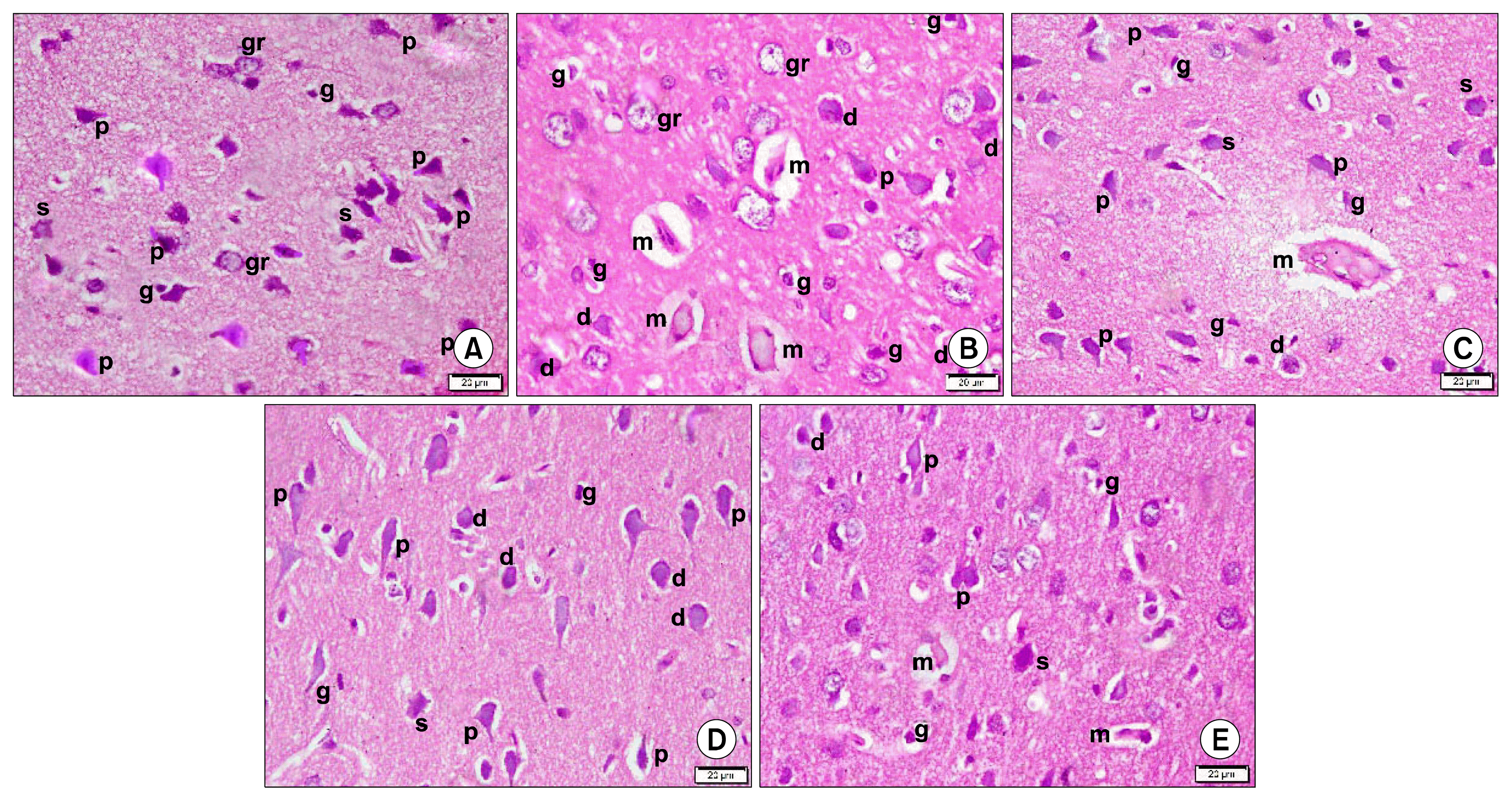 | Fig. 1Photomicrographs of sections in the frontal area of cerebral cortex (EPL) (H&E, ×400). (A) of a rat in group 1 showing multiple pyramidal (p), few stellate (s) and few granule (gr) neurons. Note few glial cells (g) in neuropil. (B) of a rat in group 2 showing 4 acidophilic masses (m) exhibiting dark nuclei and surrounded by a clear space. Multiple deformed (d), few pyramidal (p) and granule (gr) neurons are seen. Note multiple glial cells (g). (C) of a rat in group 3 showing an acidophilic mass (m) exhibiting dark nuclei and surrounded by a clear space. Few deformed (d), some pyramidal (p) and few stellate (s) neurons are seen. Note some glial cells (g). (D) of a rat in subgroup 4a showing few deformed (d), multiple pyramidal (p) and few stellate (s) neurons. Note few glial cells (g). (E) of a rat in subgroup 4b showing 2 small acidophilic masses (m) exhibiting dark nuclei and surrounded by a clear space. Few deformed (d), multiple pyramidal (p) and few stellate (s) neurons are seen. Note few glial cells (g). |
Histochemical results
 | Fig. 2Photomicrographs of sections in the frontal area of cerebral cortex (EPL) (CR ×200). (A) of a rat in group 1 showing dull Congo red (CR) staining of neurons (N) and neuropil (n). (B) of a rat in group 2 showing 4 CR +ve masses (arrowheads). (C) of a rat in group 3 showing 3 small CR +ve masses (arrowheads). (D) of a rat in subgroup 4a showing dull Congo red (CR) staining of neurons (N) and neuropil (n). Note multiple blood vessels (v). (E) of a rat in subgroup 4b showing a small CR+ve mass (arrowhead) (CR ×200). |
Immunohistochemical results
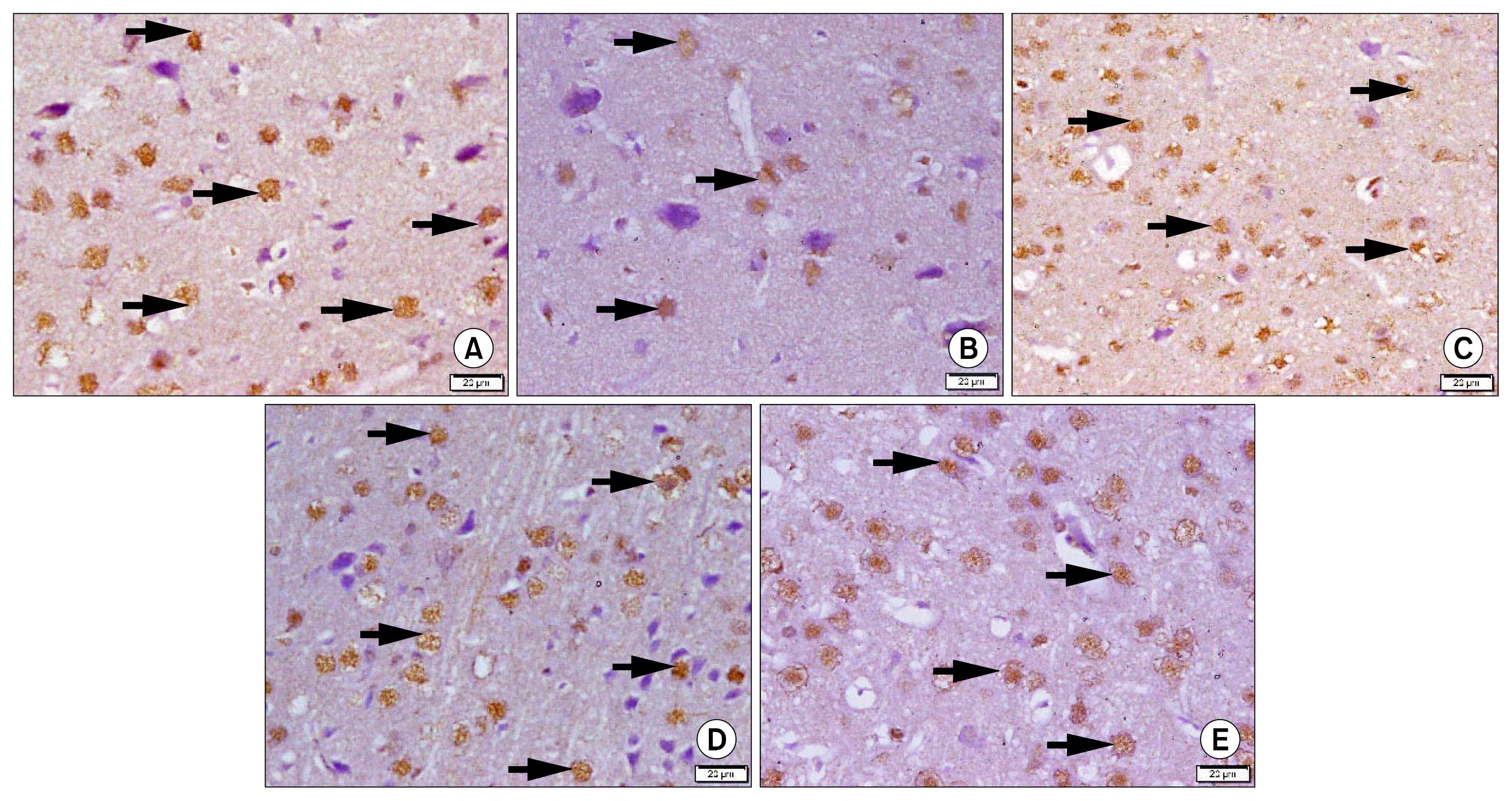 | Fig. 3Photomicrographs of sections in the frontal area of cerebral cortex (EPL) (P-CREB immuno-staining, ×400). (A) of a rat in group 1 showing multiple +ve nuclei of neurons (arrows). (B) of a rat in group 2 showing few +ve nuclei of neurons (arrows). (C) of a rat in group 3 showing some +ve nuclei of neurons (arrows). (D) of a rat in subgroup 4a showing multiple +ve nuclei of neurons (arrows). (E) of a rat in subgroup 4b showing multiple +ve nuclei of neurons (arrows). |
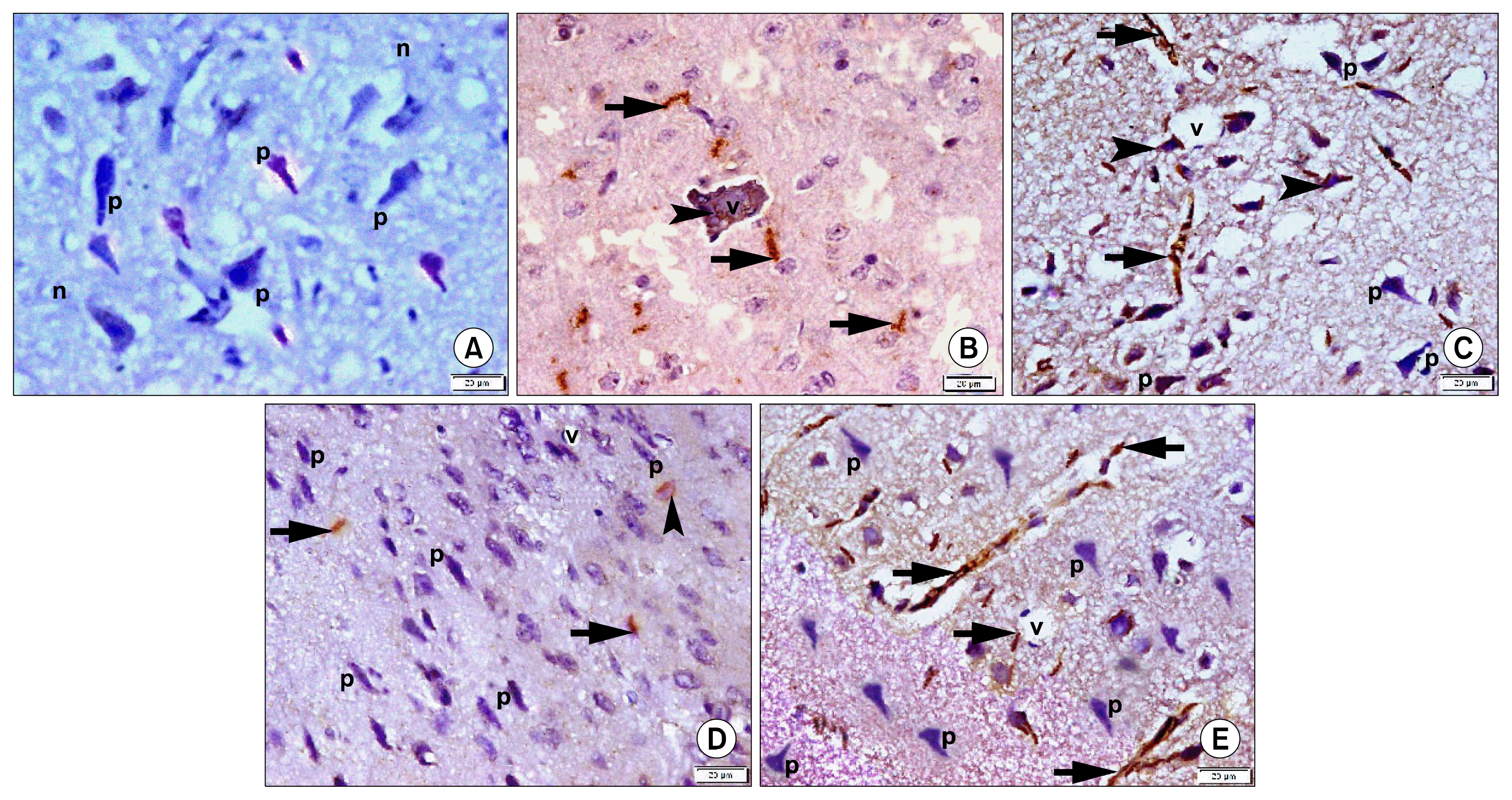 | Fig. 4Photomicrographs of sections in the frontal area of cerebral cortex (EPL) (CD44 immuno-staining, ×400). (A) of a rat in group 1 showing −ve immunostaining of pyramidal neurons (p) and neuropil (n). (B) of a rat in group 2 showing some +ve spindle cells around (arrows) and inside (arrowhead) a blood vessel (v). (C) of a rat in group 3 showing multiple +ve cells in neuropil (arrows) and fused with neurons (arrowheads). Note some pyramidal neurons (p). (D) of a rat in subgroup 4a showing few +ve cells in neuropil (arrows) and fused with a neuron (arrowhead). Note multiple pyramidal neurons (p) and a vessel (v). (E) of a rat in subgroup 4b showing multiple +ve cells in neuropil (arrows) and multiple pyramidal neurons (p). |
Morphometric results
Table 1
| Group | Area of deformed | Area of glial cells | Area of plaques | Area % of | Area % of |
|---|---|---|---|---|---|
| neurons | +ve P-CREB | +ve CD44 | |||
| Group 1 | - | 0.76±0.09 | - | 25.01±6.52 | - |
| Group 2 | 7.54±1.72 | 3.81±0.51# | 733.16±31.23 | 8.07±1.62$ | 2.98±0.53 |
| Group 3 | 3.31±0.49* | 0.94±0.12 | 148.44±18.01^ | 17.39±4.08 | 6.44±0.92@ |
| Subgroup 4a | 1.98±0.12** | 0.74±0.05 | - | 26.08±3.22 | 1.67±0.21 |
| Subgroup 4b | 1.81±0.05** | 0.80±0.20 | 68.74±5.94^^ | 20.75±4.22 | 7.36±1.05@ |




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download