2. Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, O'Sullivan C, Collins L, Chen Y, Minami E, Gill EA, Ueno S, Yuan C, Gold J, Murry CE. 2007; Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol. 25:1015–1024. DOI:
10.1038/nbt1327. PMID:
17721512.

3. Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, Hsiao C, Kamp TJ, Palecek SP. 2013; Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat Protoc. 8:162–175. DOI:
10.1038/nprot.2012.150. PMID:
23257984. PMCID:
PMC3612968.

4. Cao N, Liang H, Huang J, Wang J, Chen Y, Chen Z, Yang HT. 2013; Highly efficient induction and long-term maintenance of multipotent cardiovascular progenitors from human pluripotent stem cells under defined conditions. Cell Res. 23:1119–1132. DOI:
10.1038/cr.2013.102. PMID:
23896987. PMCID:
PMC3773575.

5. Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, Lan F, Diecke S, Huber B, Mordwinkin NM, Plews JR, Abilez OJ, Cui B, Gold JD, Wu JC. 2014; Chemically defined generation of human cardiomyocytes. Nat Methods. 11:855–860. DOI:
10.1038/nmeth.2999. PMID:
24930130. PMCID:
PMC4169698.

6. Melkoumian Z, Weber JL, Weber DM, Fadeev AG, Zhou Y, Dolley-Sonneville P, Yang J, Qiu L, Priest CA, Shogbon C, Martin AW, Nelson J, West P, Beltzer JP, Pal S, Brandenberger R. 2010; Synthetic peptide-acrylate surfaces for long-term self-renewal and cardiomyocyte differentiation of human embryonic stem cells. Nat Biotechnol. 28:606–610. DOI:
10.1038/nbt.1629. PMID:
20512120.

7. Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ, Palecek SP. 2012; Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci U S A. 109:E1848–E1857. DOI:
10.1073/pnas.1200250109. PMID:
22645348. PMCID:
PMC3390875.

8. Schmid P, Lorenz A, Hameister H, Montenarh M. 1991; Expression of p53 during mouse embryogenesis. Development. 113:857–865. DOI:
10.1242/dev.113.3.857. PMID:
1821855.

9. Lutzker SG, Levine AJ. 1996; A functionally inactive p53 protein in teratocarcinoma cells is activated by either DNA damage or cellular differentiation. Nat Med. 2:804–810. DOI:
10.1038/nm0796-804. PMID:
8673928.

10. Wallingford JB, Seufert DW, Virta VC, Vize PD. 1997; p53 activity is essential for normal development in Xenopus. Curr Biol. 7:747–757. DOI:
10.1016/S0960-9822(06)00333-2. PMID:
9368757.

11. Shigeta M, Ohtsuka S, Nishikawa-Torikai S, Yamane M, Fujii S, Murakami K, Niwa H. 2013; Maintenance of pluripotency in mouse ES cells without Trp53. Sci Rep. 3:2944. DOI:
10.1038/srep02944. PMID:
24126347. PMCID:
PMC3796736.

12. Wang Q, Zou Y, Nowotschin S, Kim SY, Li QV, Soh CL, Su J, Zhang C, Shu W, Xi Q, Huangfu D, Hadjantonakis AK, Massagué J. 2017; The p53 family coordinates Wnt and nodal inputs in mesendodermal differentiation of embryonic stem cells. Cell Stem Cell. 20:70–86. DOI:
10.1016/j.stem.2016.10.002. PMID:
27889317. PMCID:
PMC5218926.

13. Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Izpisúa Belmonte JC. 2009; Linking the p53 tumour suppressor pathway to somatic cell repro-gramming. Nature. 460:1140–1144. DOI:
10.1038/nature08311. PMID:
19668186. PMCID:
PMC2735889.

14. Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S. 2009; Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 460:1132–1135. DOI:
10.1038/nature08235. PMID:
19668191. PMCID:
PMC2917235.

15. Zhu J, Dou Z, Sammons MA, Levine AJ, Berger SL. 2016; Lysine methylation represses p53 activity in teratocarcinoma cancer cells. Proc Natl Acad Sci U S A. 113:9822–9827. DOI:
10.1073/pnas.1610387113. PMID:
27535933. PMCID:
PMC5024588.

16. Cordenonsi M, Dupont S, Maretto S, Insinga A, Imbriano C, Piccolo S. 2003; Links between tumor suppressors: p53 is required for TGF-beta gene responses by cooperating with Smads. Cell. 113:301–314. DOI:
10.1016/S0092-8674(03)00308-8. PMID:
12732139.
17. Lee KH, Li M, Michalowski AM, Zhang X, Liao H, Chen L, Xu Y, Wu X, Huang J. 2010; A genomewide study identifies the Wnt signaling pathway as a major target of p53 in murine embryonic stem cells. Proc Natl Acad Sci U S A. 107:69–74. DOI:
10.1073/pnas.0909734107. PMID:
20018659. PMCID:
PMC2806696.

18. Zhou R, Huang W, Fan X, Liu F, Luo L, Yuan H, Jiang Y, Xiao H, Zhou Z, Deng C, Dang X. 2019; miR-499 released during myocardial infarction causes endothelial injury by targeting α7-nAchR. J Cell Mol Med. 23:6085–6097. DOI:
10.1111/jcmm.14474. PMID:
31270949. PMCID:
PMC6714230.

19. Liu Y, Elf SE, Miyata Y, Sashida G, Liu Y, Huang G, Di Giandomenico S, Lee JM, Deblasio A, Menendez S, Antipin J, Reva B, Koff A, Nimer SD. 2009; p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell. 4:37–48. DOI:
10.1016/j.stem.2008.11.006. PMID:
19128791. PMCID:
PMC2839936.

20. Meletis K, Wirta V, Hede SM, Nistér M, Lundeberg J, Frisén J. 2006; p53 suppresses the self-renewal of adult neural stem cells. Development. 133:363–369. DOI:
10.1242/dev.02208. PMID:
16368933.

21. Jain AK, Allton K, Iacovino M, Mahen E, Milczarek RJ, Zwaka TP, Kyba M, Barton MC. 2012; p53 regulates cell cycle and microRNAs to promote differentiation of human embryonic stem cells. PLoS Biol. 10:e1001268. DOI:
10.1371/journal.pbio.1001268. PMID:
22389628. PMCID:
PMC3289600.

22. Li M, He Y, Dubois W, Wu X, Shi J, Huang J. 2012; Distinct regulatory mechanisms and functions for p53-activated and p53-repressed DNA damage response genes in embryonic stem cells. Mol Cell. 46:30–42. DOI:
10.1016/j.molcel.2012.01.020. PMID:
22387025. PMCID:
PMC3327774.

23. Akdemir KC, Jain AK, Allton K, Aronow B, Xu X, Cooney AJ, Li W, Barton MC. 2014; Genome-wide profiling reveals stimulus-specific functions of p53 during differentiation and DNA damage of human embryonic stem cells. Nucleic Acids Res. 42:205–223. DOI:
10.1093/nar/gkt866. PMID:
24078252. PMCID:
PMC3874181.

24. Cordenonsi M, Montagner M, Adorno M, Zacchigna L, Martello G, Mamidi A, Soligo S, Dupont S, Piccolo S. 2007; Integration of TGF-beta and Ras/MAPK signaling through p53 phosphorylation. Science. 315:840–843. DOI:
10.1126/science.1135961. PMID:
17234915.

25. Foulquier S, Daskalopoulos EP, Lluri G, Hermans KCM, Deb A, Blankesteijn WM. 2018; WNT signaling in cardiac and vascular disease. Pharmacol Rev. 70:68–141. DOI:
10.1124/pr.117.013896. PMID:
29247129. PMCID:
PMC6040091.

26. Zhao M, Tang Y, Zhou Y, Zhang J. 2019; Deciphering role of Wnt signalling in cardiac mesoderm and cardiomyocyte differentiation from human iPSCs: four-dimensional control of Wnt pathway for hiPSC-CMs differentiation. Sci Rep. 9:19389. DOI:
10.1038/s41598-019-55620-x. PMID:
31852937. PMCID:
PMC6920374.

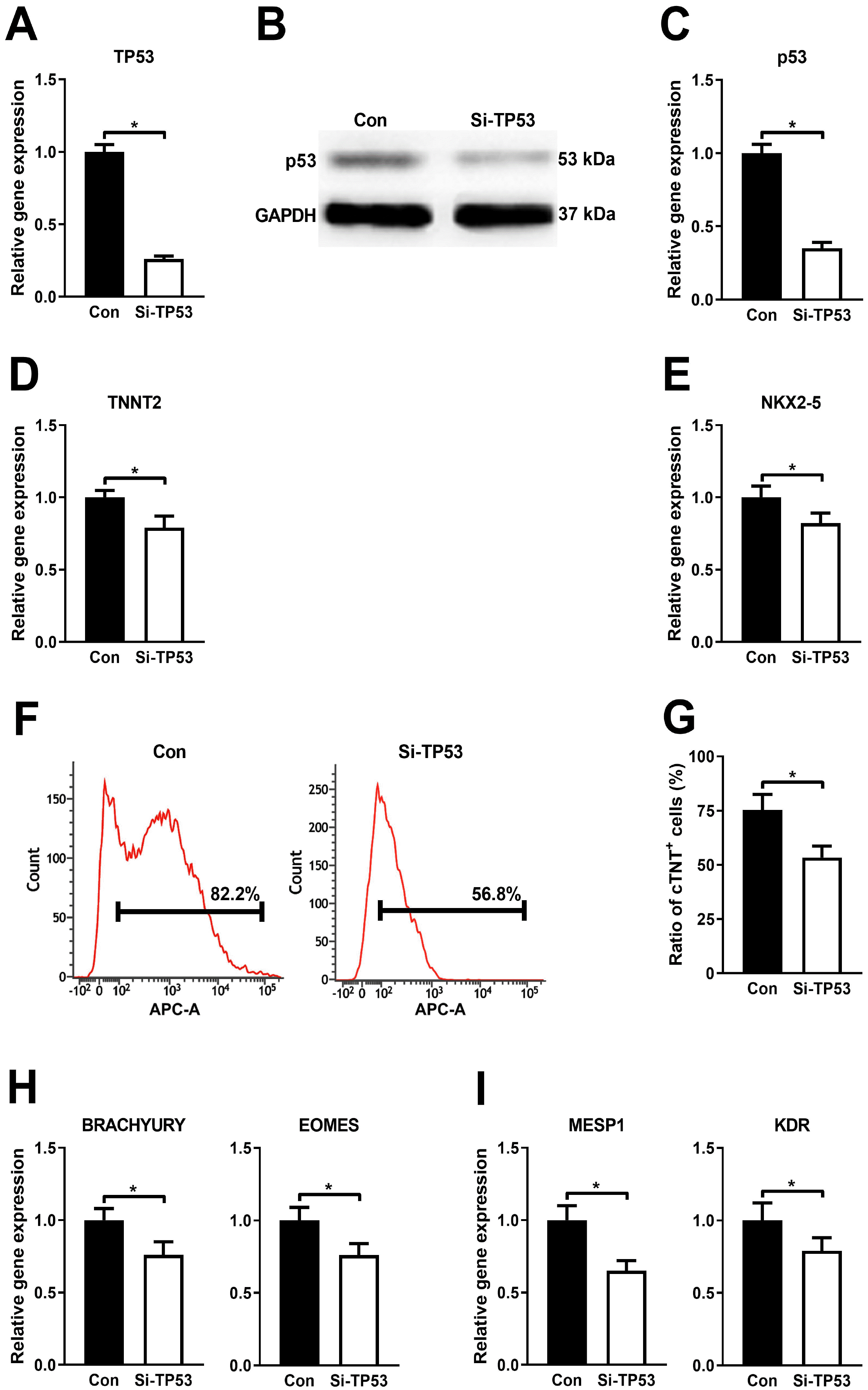
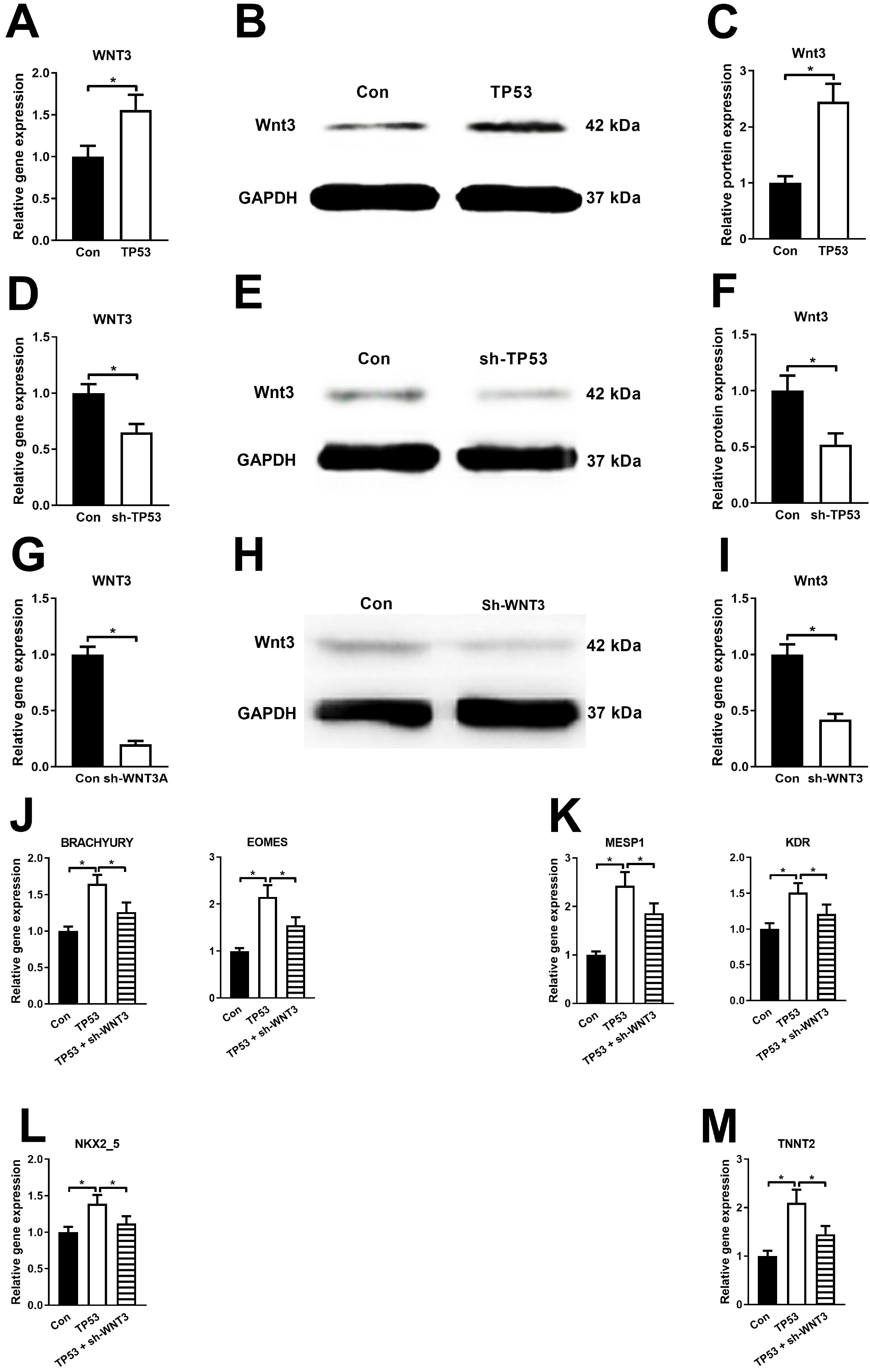
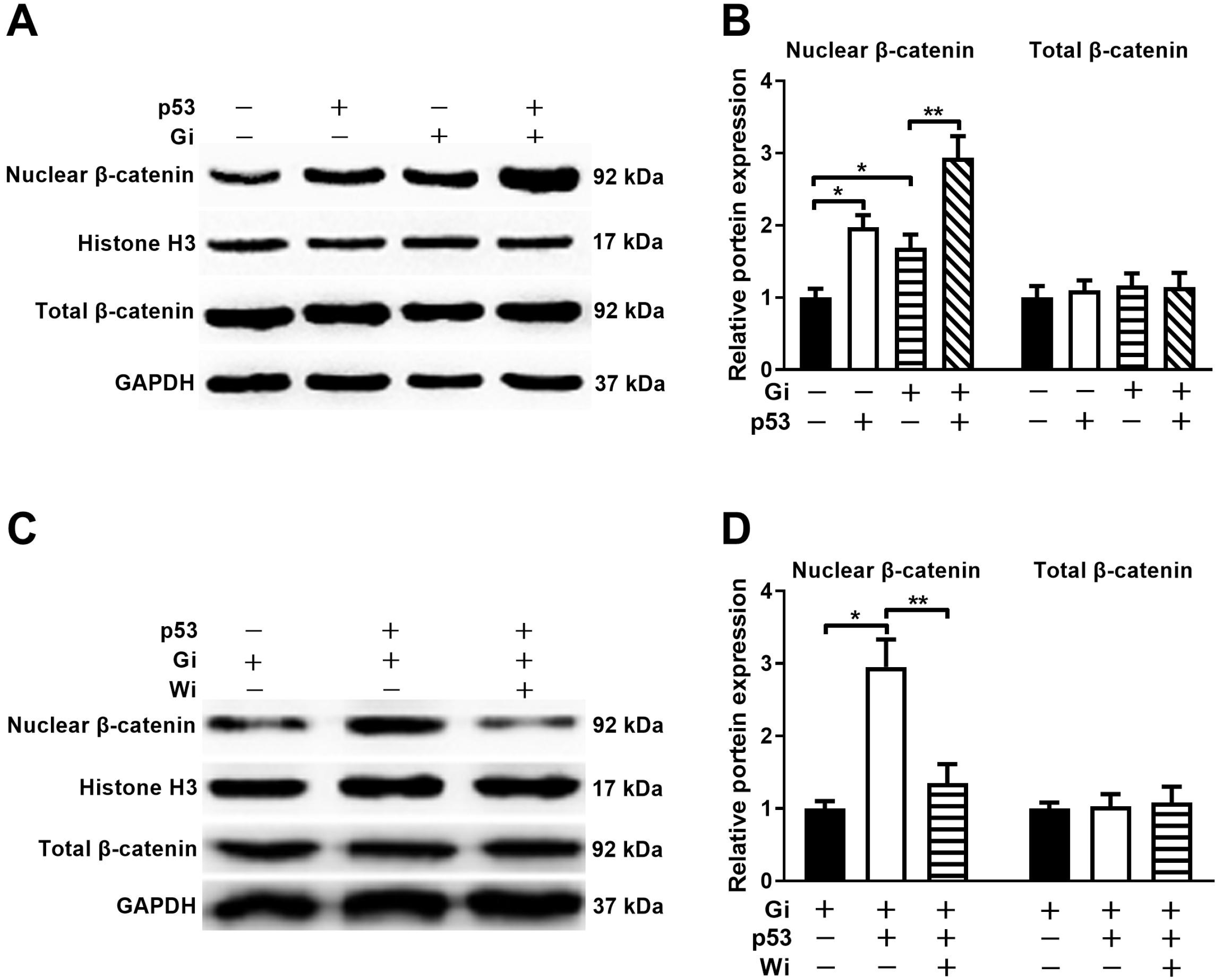




 PDF
PDF Citation
Citation Print
Print


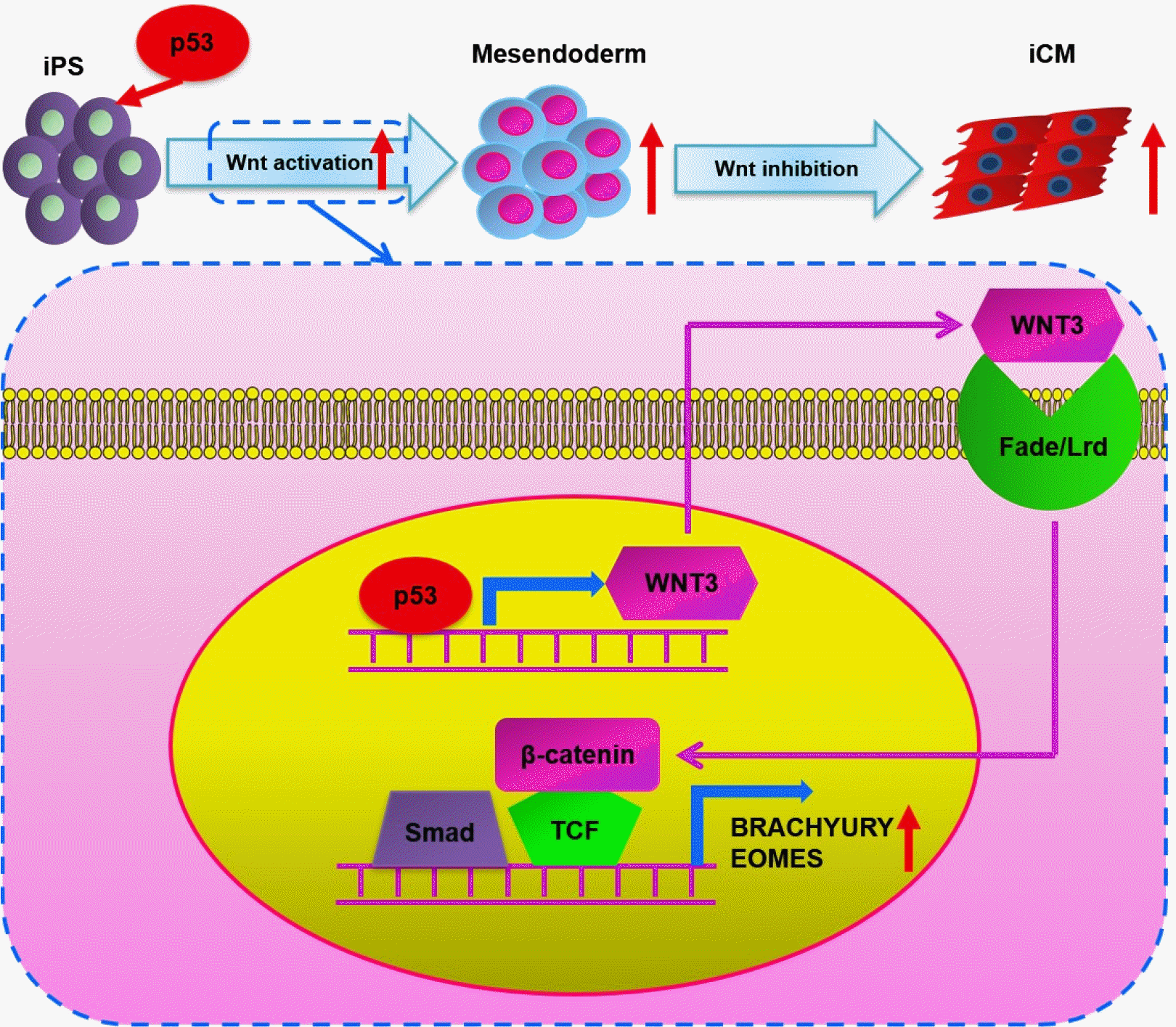
 XML Download
XML Download