Introduction
Materials and Methods
Ethics statement
Establishment rabbit model of lumbar fusion
Bioinformatics analysis
Cell culture and differentiation
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Table 1
Enzyme linked immuno sorbent assay (ELISA)
Alizarin red staining
Cell transfection
Dual luciferase reporter gene assay
Cell counting kit (CCK)-8
Western blot
Statistical analysis
Results
MiR-34c-5p was positively correlated with osteogenic differentiation
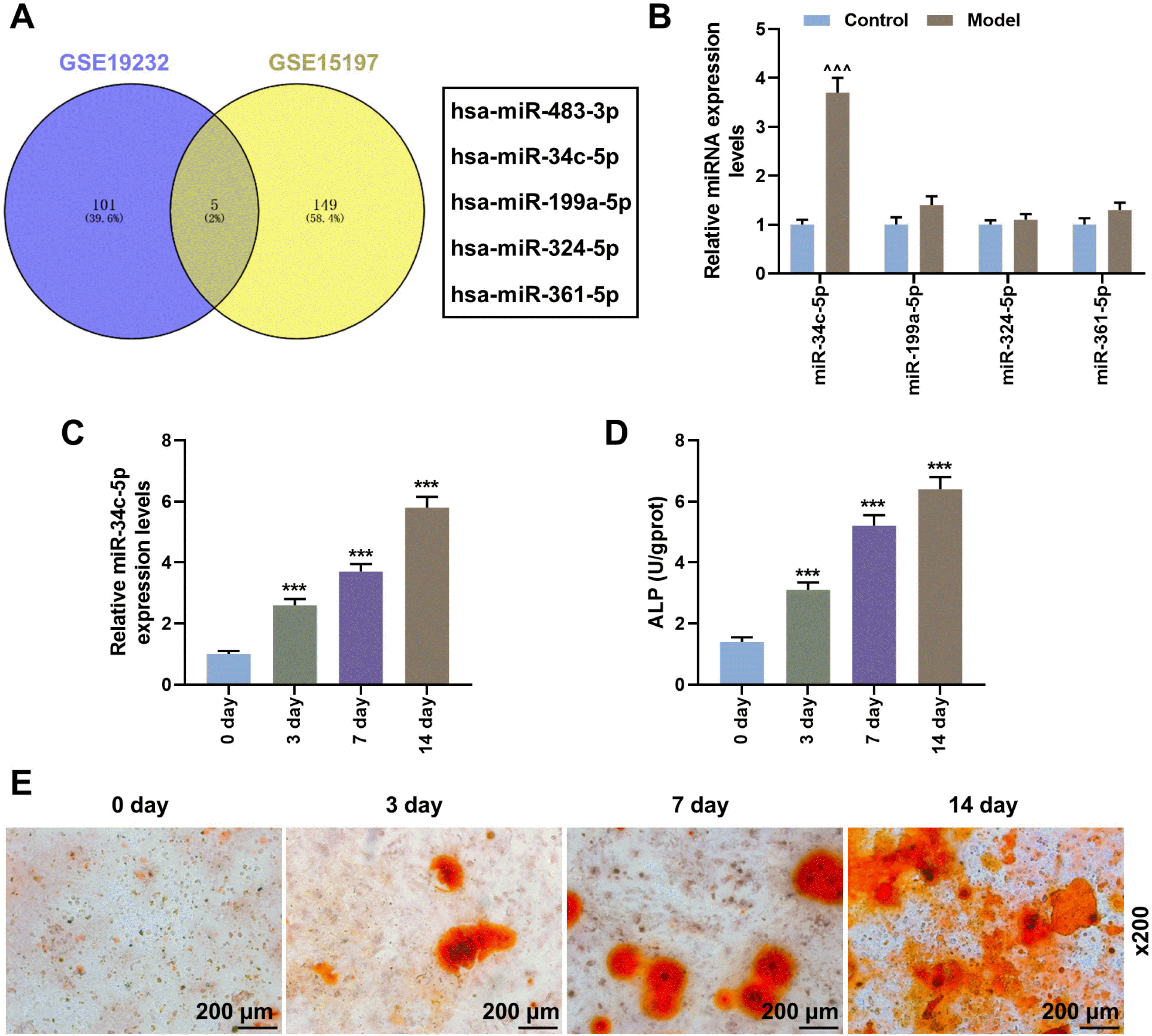 | Fig. 1During osteogenic differentiation of BMSCs, miR-34c-5p expression, ALP content and calcium deposition were promoted. (A) Abnormal expressions of miRNAs during osteogenic differentiation of mesenchymal stem cells obtained from the GEO database (GSE115197 and GSE19232). (B) The expressions of miR-34c-5p, miR-199a-5p, miR-324-5p and miR-361-5p in rabbit lumbar fusion model rabbit or normal rabbit were determined by qRT-PCR. (C) qRT-PCR was used to detect the expression of miR-34c-5p in BMSCs with different differentiation times (0, 3, 7 and 14 day). (D) ELISA was used to measure the content of ALP in BMSCs at different differentiation times (0, 3, 7 and 14 day). (E) Alizarin red staining of BMSCs at different differentiation times (0, 3, 7 and 14 day). Scales: 200 μm; magnifications: ×200. ^vs. Control, *vs. 0 days. ^^^ or ***p<0.001. BMSCs, bone marrow mesenchymal stem cells; ALP, alkaline phosphatase; GEO, gene expression omnibus; qRT-PCR, quantitative reverse transcription polymerase chain reaction; ELISA, enzyme linked immune sorbent assay. The experiment was independently repeated three times. |
MiR-34c-5p promoted osteogenic differentiation of BMSCs
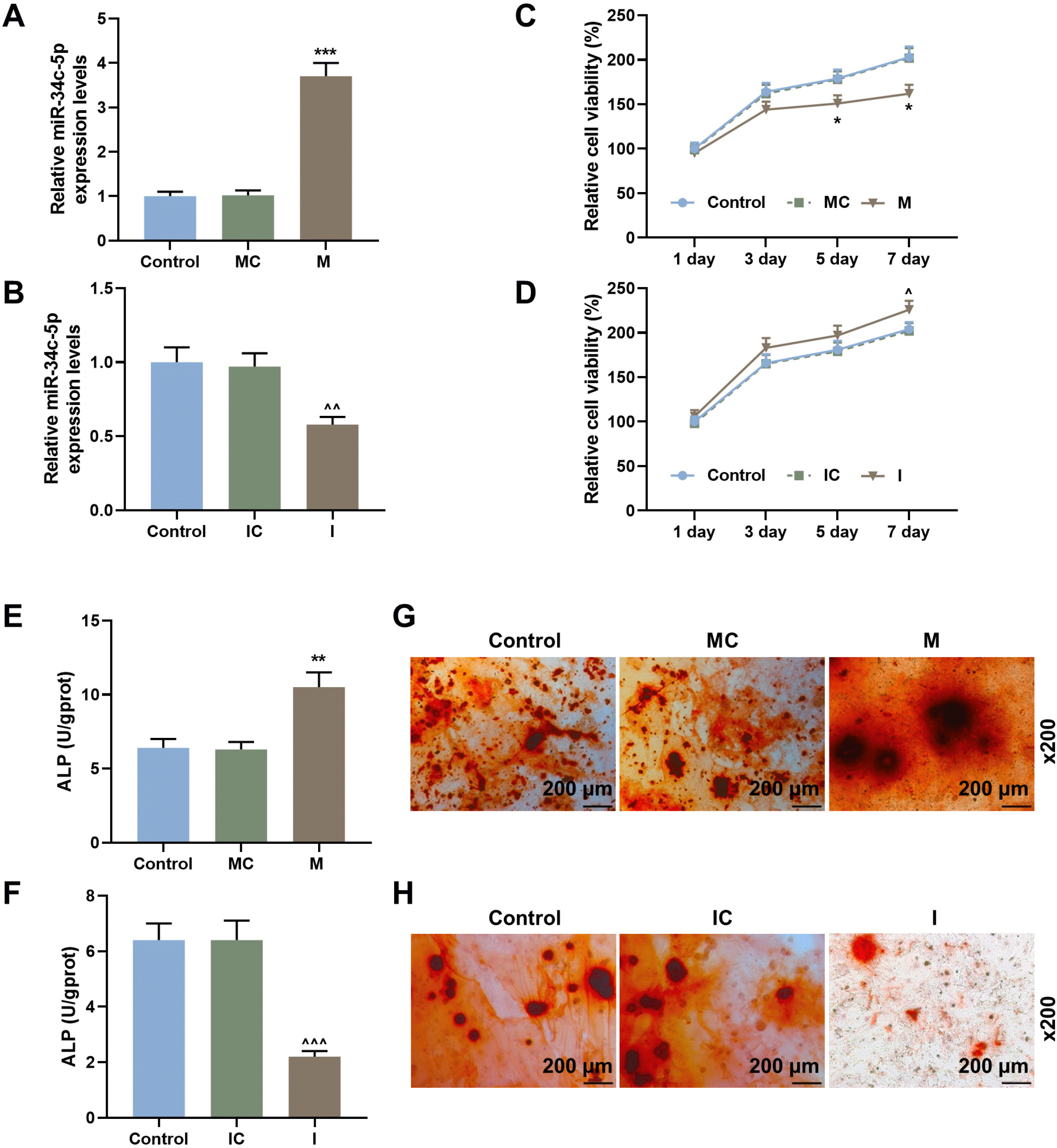 | Fig. 2miR-34c-5p decreased cell proliferation and increased ALP content and calcium deposits of BMSCs. (A) The expression of miR-34c-5p in BMSCs transfected with miR-34c-5p mimic or mimic control was detected by qRT-PCR. (B) The expression of miR-34c-5p in BMSCs transfected with miR-34c-5p inhibitor or inhibitor control was detected by qRT-PCR. (C) The cell proliferation of BMSCs transfected with miR-34c-5p mimic or mimic control was identified by CCK-8 at different times (1, 3, 5 and 7 day). (D) The cell proliferation of BMSCs transfected with miR-34c-5p inhibitor or inhibitor control was identified by CCK-8 at different times (1, 3, 5 and 7 day). (E) The ALP content of BMSCs transfected with miR-34c-5p mimic or mimic control was detected by ELISA. (F) The ALP content of BMSCs transfected with miR-34c-5p inhibitor or inhibitor control was detected by ELISA. (G) Alizarin red staining of BMSCs transfected with miR-34c-5p mimic or mimic control at 14 days. Scales: 200 μm; magnifications: ×200. (H) Alizarin red staining of BMSCs transfected with miR-34c-5p inhibitor or inhibitor control at 14 days. Scales: 200 μm; magnifications: ×200. *vs. MC; ^vs. IC. * or ^p<0.05; ** or ^^p<0.01; *** or ^^^p<0.001. BMSCs, bone marrow mesenchymal stem cells; qRT-PCR, quantitative reverse transcription polymerase chain reaction; CCK-8, cell counting 8; ALP, alkaline phosphatase; ELISA, enzyme linked immune sorbent assay; I, miR-34c-5p inhibitor; IC, inhibitor control; M, miR-34c-5p mimic; MC, mimic control. The experiment was independently repeated three times. |
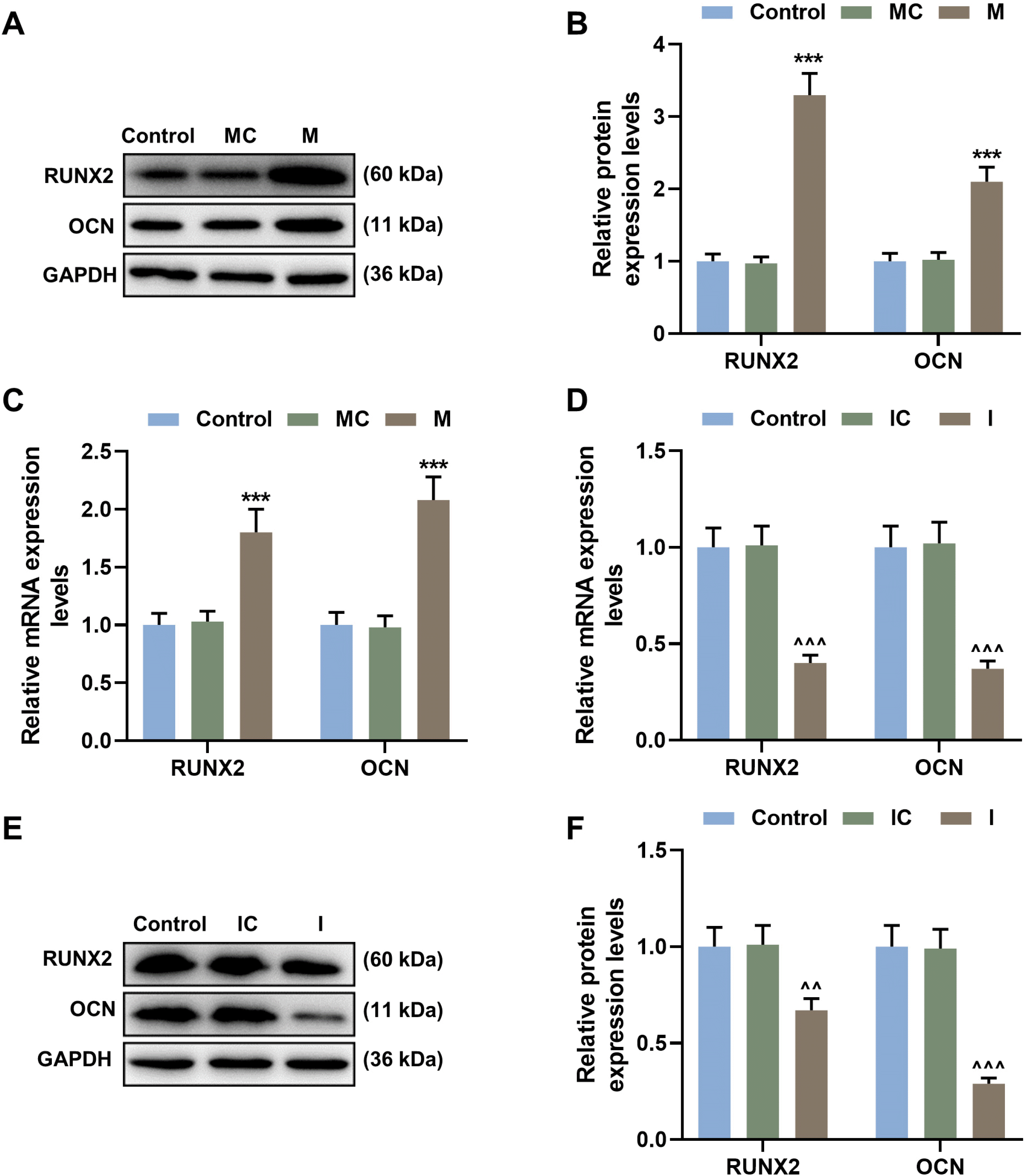 | Fig. 3MiR-34c-5p promoted the expression of RUNX2 and OCN of BMSCs. (A, B) The expression of RUNX2 and OCN of BMSCs transfected with miR-34c-5p mimic or mimic control was detected by Western blot. (C) The expressions of RUNX2 and OCN of BMSCs transfected with miR-34c-5p mimic or mimic control were detected by qRT-PCR. (D, E) The expressions of RUNX2 and OCN of BMSCs transfected with miR-34c-5p inhibitor or inhibitor control were detected by Western blot. (F) The expressions of RUNX2 and OCN of BMSCs transfected with miR-34c-5p inhibitor or inhibitor control were detected by qRT-PCR. *vs. MC; ^vs. IC. ^^p<0.01; *** or ^^^p<0.001. BMSCs, bone marrow mesenchymal stem cells; RUNX2, Runt-related transcription factor 2; OCN, osteocalcin; qRT-PCR, quantitative reverse transcription polymerase chain reaction; I, miR-34c-5p inhibitor; IC, inhibitor control; M, miR-34c-5p mimic; MC, mimic control. The experiment was independently repeated three times. |
MiR-34c-5p promoted osteogenic differentiation of BMSCs by inhibiting the expression of Bcl-2
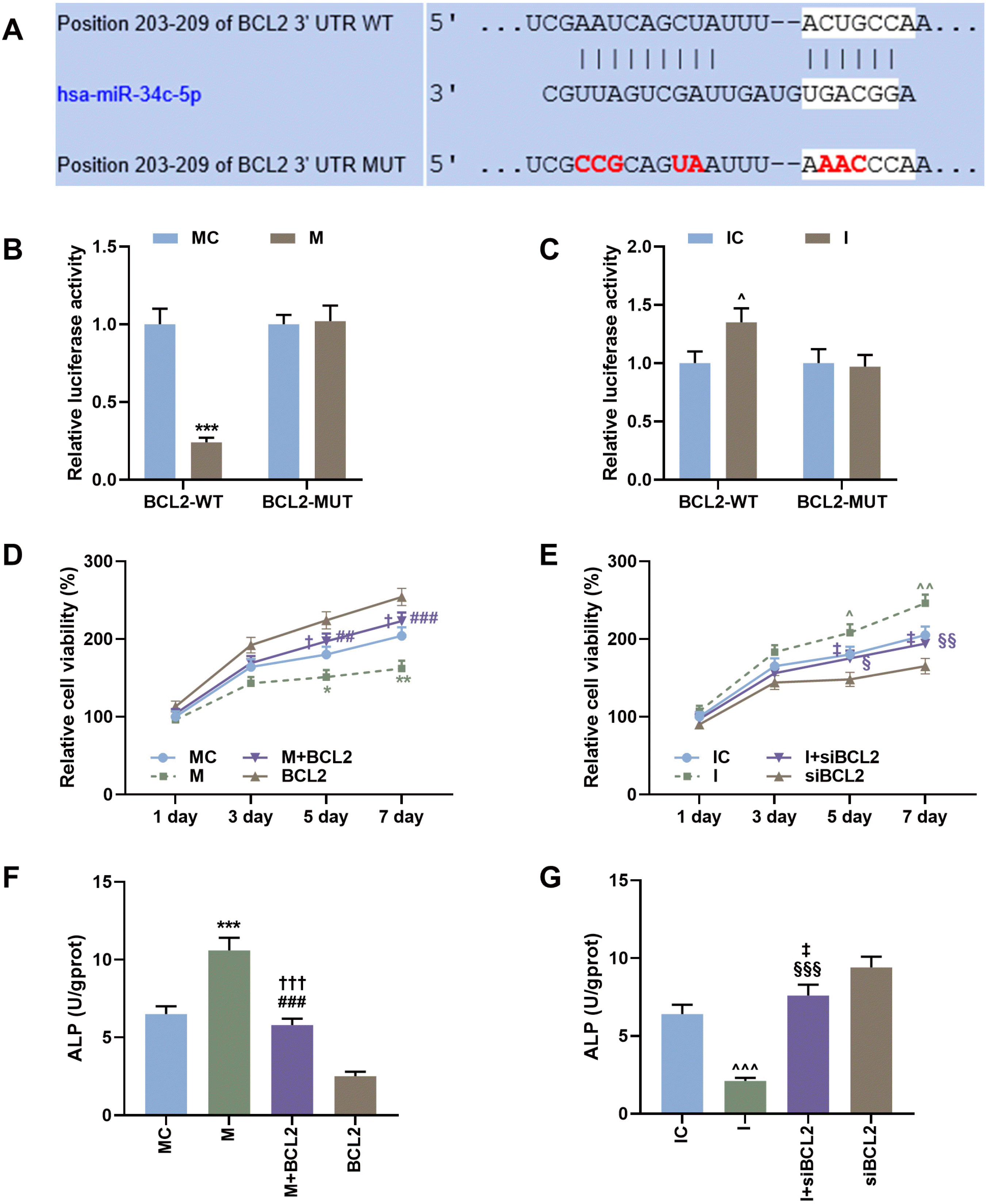 | Fig. 4MiR-34c-5p decreased cell activity and increased ALP content of BMSCs by inhibiting Bcl-2 expression. (A) The binding site of miR-34c-5p and Bcl-2 was predicted by TargetScan. (B, C) Dual-luciferase reporter gene assay was used to verify the targeted binding relationship between miR-34c-5p and Bcl-2. (D) The cell proliferation of BMSCs transfected with or untransfected miR-34c-5p mimic and/or Bcl-2 was detected by CCK-8 at 1, 3, 5 and 7 day. (E) The cell proliferation of BMSCs treated or untreated with miR-34c-5p inhibitor and/or siBcl-2 was detected by CCK-8 at 1, 3, 5 and 7 day. (F) The ALP content of BMSCs treated or untreated with miR-34c-5p mimic and/or Bcl-2 was detected by ELISA. (G) The ALP content of BMSCs treated or untreated with miR-34c-5p inhibitor and/or siBcl-2 was detected by ELISA. *vs. MC, #vs. M, †vs. BCL2, ^vs. IC, §vs. I, ‡vs. siBCL2. † or ^ or ‡ or §p<0.05; ## or ^^ or §§p<0.01; *** or ^^^ or ††† or ###p<0.001. BMSCs, bone marrow mesenchymal stem cells; CCK-8, cell counting 8; ALP, alkaline phosphatase; ELISA, enzyme linked immune sorbent assay; Bcl-2, B cell lymphoma/lewkmia-2; I, miR-34c-5p inhibitor; IC, inhibitor control; M, miR-34c-5p mimic; MC, mimic control. The experiment was independently repeated three times. |
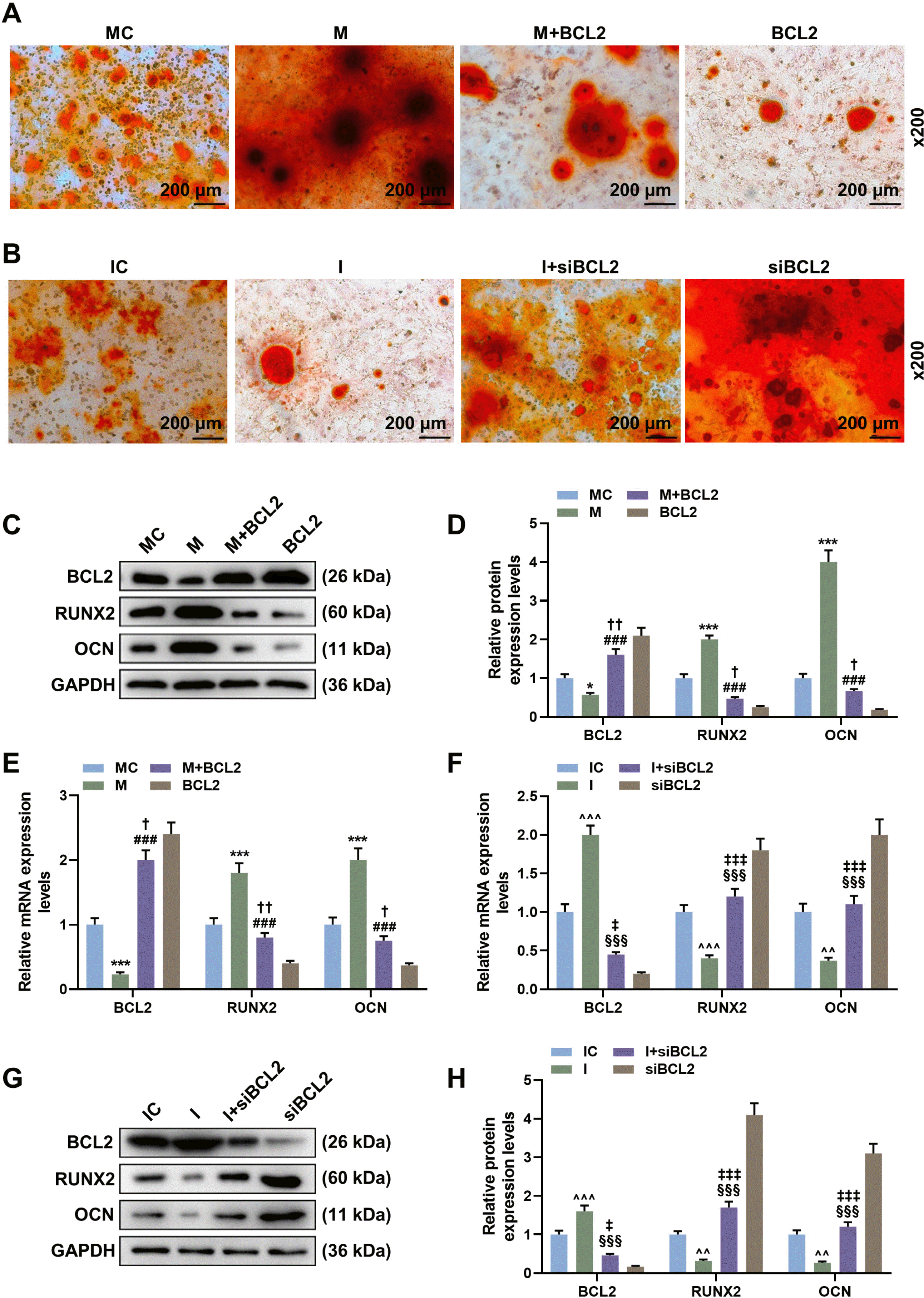 | Fig. 5MiR-34c-5p promoted calcium deposits and the expression of RUNX2 and OCN of BMSCs by inhibiting Bcl-2 expression. (A) Alizarin red staining of BMSCs treated or untreated with miR-34c-5p mimic and/or Bcl-2 at 14 days. Scales: 200 μm; magnifications: ×200. (B) Alizarin red staining of BMSCs treated or untreated with miR-34c-5p inhibitor and/or siBcl-2 at 14 days. Scales: 200 μm; magnifications: ×200. (C, D) The expression of Bcl-2, RUNX2 and OCN of BMSCs treated or untreated with miR-34c-5p mimic and/or Bcl-2 were detected by Western blot. (E) The expressions of Bcl-2, RUNX2 and OCN of BMSCs treated or untreated with miR-34c-5p mimic and/or Bcl-2 were detected by qRT-PCR. (F) The expressions of Bcl-2, RUNX2 and OCN of BMSCs treated or untreated with miR-34c-5p inhibitor and/or siBcl-2 were detected by qRT-PCR. (G, H) The expressions of Bcl-2, RUNX2 and OCN of BMSCs treated or untreated with miR-34c-5p inhibitor and/or siBcl-2 were detected by Western blot. *vs. MC, #vs. M, †vs. BCL2, ^vs. IC, §vs. I, ‡vs. siBCL2. † or * or ‡p<0.05; ^^ or ††p<0.01; *** or ^^^ or ‡‡‡ or ### or §§§p<0.001. BMSCs, bone marrow mesenchymal stem cells; CCK-8, cell counting 8; ALP, alkaline phosphatase; ELISA, enzyme linked immune sorbent assay; Bcl-2, B cell lymphoma/lewkmia-2; I, miR-34c-5p inhibitor; IC, inhibitor control; M, miR-34c-5p mimic; MC, mimic control. The experiment was independently repeated three times. |




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download