1. Abate-Shen C, Shen MM, Gelmann E. 2008; Integrating differentiation and cancer: the Nkx3.1 homeobox gene in prostate organogenesis and carcinogenesis. Differentiation. 76:717–727. DOI:
10.1111/j.1432-0436.2008.00292.x. PMID:
18557759. PMCID:
PMC3683569.

3. Bieberich CJ, Fujita K, He WW, Jay G. 1996; Prostate-specific and androgen-dependent expression of a novel homeobox gene. J Biol Chem. 271:31779–31782. DOI:
10.1074/jbc.271.50.31779. PMID:
8943214.

4. Abdulkadir SA, Magee JA, Peters TJ, Kaleem Z, Naughton CK, Humphrey PA, Milbrandt J. 2002; Conditional loss of Nkx3.1 in adult mice induces prostatic intraepithelial neoplasia. Mol Cell Biol. 22:1495–1503. DOI:
10.1128/MCB.22.5.1495-1503.2002. PMID:
11839815. PMCID:
PMC134699.

5. Sfanos KS, Yegnasubramanian S, Nelson WG, De Marzo AM. 2018; The inflammatory microenvironment and microbiome in prostate cancer development. Nat Rev Urol. 15:11–24. DOI:
10.1038/nrurol.2017.167. PMID:
29089606.

6. Cai T, Santi R, Tamanini I, Galli IC, Perletti G, Bjerklund Johansen TE, Nesi G. 2019; Current knowledge of the potential links between inflammation and prostate cancer. Int J Mol Sci. 20:3833. DOI:
10.3390/ijms20153833. PMID:
31390729. PMCID:
PMC6696519.

7. McArdle PA, Canna K, McMillan DC, McNicol AM, Campbell R, Underwood MA. 2004; The relationship between T-lymphocyte subset infiltration and survival in patients with prostate cancer. Br J Cancer. 91:541–543. DOI:
10.1038/sj.bjc.6601943. PMID:
15266325. PMCID:
PMC2409839.

8. Nonomura N, Takayama H, Nakayama M, Nakai Y, Kawashima A, Mukai M, Nagahara A, Aozasa K, Tsujimura A. 2011; Infiltration of tumour-associated macrophages in prostate biopsy specimens is predictive of disease progression after hormonal therapy for prostate cancer. BJU Int. 107:1918–1922. DOI:
10.1111/j.1464-410X.2010.09804.x. PMID:
21044246.

10. Keller ET, Ershler WB, Chang C. 1996; The androgen receptor: a mediator of diverse responses. Front Biosci. 1:d59–d71. DOI:
10.2741/A116. PMID:
9159212.

12. Tan PY, Chang CW, Chng KR, Wansa KD, Sung WK, Cheung E. 2012; Integration of regulatory networks by NKX3-1 promotes androgen-dependent prostate cancer survival. Mol Cell Biol. 32:399–414. DOI:
10.1128/MCB.05958-11. PMID:
22083957. PMCID:
PMC3255774.

13. Kim KP, Wu Y, Yoon J, Adachi K, Wu G, Velychko S, MacCarthy CM, Shin B, Röpke A, Arauzo-Bravo MJ, Stehling M, Han DW, Gao Y, Kim J, Gao S, Schöler HR. 2020; Reprogramming competence of OCT factors is determined by transactivation domains. Sci Adv. 6:eaaz7364. DOI:
10.1126/sciadv.aaz7364. PMID:
32917606. PMCID:
PMC7467702.

14. Padmanabhan A, Rao V, De Marzo AM, Bieberich CJ. 2016; Regulating NKX3.1 stability and function: post-translational modifications and structural determinants. Prostate. 76:523–533. DOI:
10.1002/pros.23144. PMID:
26841725.

15. Thomas MA, Preece DM, Bentel JM. 2010; Androgen regulation of the prostatic tumour suppressor NKX3.1 is mediated by its 3' untranslated region. Biochem J. 425:575–583. DOI:
10.1042/BJ20091109. PMID:
19886863.

16. Yu CX, Jin T, Chen WW, Zhang PJ, Liu WW, Guan HY, Zhang J, Liu QW, Jiang AL. 2009; Identification of Sp1-elements in the promoter region of human homeobox gene NKX3.1. Mol Biol Rep. 36:2353–2360. DOI:
10.1007/s11033-009-9457-y. PMID:
19263243.

17. Preece DM, Harvey JM, Bentel JM, Thomas MA. 2011; ETS1 regulates NKX3.1 5' promoter activity and expression in prostate cancer cells. Prostate. 71:403–414. DOI:
10.1002/pros.21254. PMID:
20842667.

18. Kruithof-de Julio M, Shibata M, Desai N, Reynon M, Halili MV, Hu YP, Price SM, Abate-Shen C, Shen MM. 2013; Canonical Wnt signaling regulates Nkx3.1 expression and luminal epithelial differentiation during prostate organogenesis. Dev Dyn. 242:1160–1171. DOI:
10.1002/dvdy.24008. PMID:
23813564. PMCID:
PMC3788082.

19. Asatiani E, Huang WX, Wang A, Rodriguez Ortner E, Cavalli LR, Haddad BR, Gelmann EP. 2005; Deletion, methylation, and expression of the NKX3.1 suppressor gene in primary human prostate cancer. Cancer Res. 65:1164–1173. DOI:
10.1158/0008-5472.CAN-04-2688. PMID:
15734999.

20. Iwata T, Schultz D, Hicks J, Hubbard GK, Mutton LN, Lotan TL, Bethel C, Lotz MT, Yegnasubramanian S, Nelson WG, Dang CV, Xu M, Anele U, Koh CM, Bieberich CJ, De Marzo AM. 2010; MYC overexpression induces prostatic intraepithelial neoplasia and loss of Nkx3.1 in mouse luminal epithelial cells. PLoS One. 5:e9427. DOI:
10.1371/journal.pone.0009427. PMID:
20195545. PMCID:
PMC2828486.

21. Ju JH, Maeng JS, Zemedkun M, Ahronovitz N, Mack JW, Ferretti JA, Gelmann EP, Gruschus JM. 2006; Physical and functional interactions between the prostate suppressor homeoprotein NKX3.1 and serum response factor. J Mol Biol. 360:989–999. DOI:
10.1016/j.jmb.2006.05.064. PMID:
16814806.

22. Zheng SL, Ju JH, Chang BL, Ortner E, Sun J, Isaacs SD, Sun J, Wiley KE, Liu W, Zemedkun M, Walsh PC, Ferretti J, Gruschus J, Isaacs WB, Gelmann EP, Xu J. 2006; Germ-line mutation of NKX3.1 cosegregates with hereditary prostate cancer and alters the homeodomain structure and function. Cancer Res. 66:69–77. DOI:
10.1158/0008-5472.CAN-05-1550. PMID:
16397218.

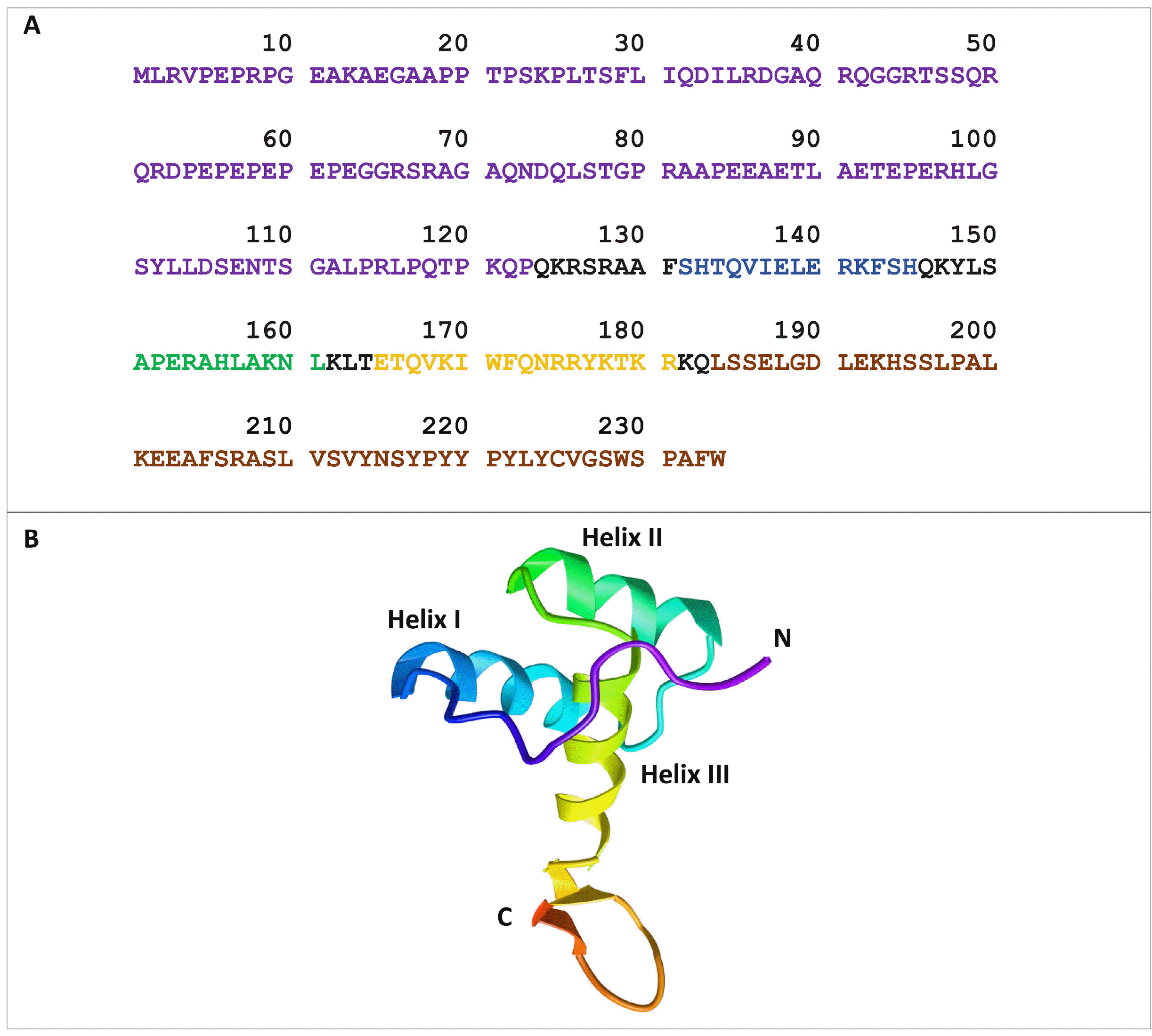
23. Liu G, Xiao R, Lee HW, Hamilton K, Ciccosanti C, Wang HB, Acton TB, Everett JK, Huang YJ, Montelione GT. 2011; Solution NMR structure of Homeobox domain of Homeobox protein Nkx-3.1 from homo sapiens, Northeast Structural Genomics Consortium Target HR6470A. doi:10.2210/pdb2L9R/pdb. DOI:
10.2210/pdb2l9r/pdb.

24. Steadman DJ, Giuffrida D, Gelmann EP. 2000; DNA-binding sequence of the human prostate-specific homeodomain protein NKX3.1. Nucleic Acids Res. 28:2389–2395. DOI:
10.1093/nar/28.12.2389. PMID:
10871372. PMCID:
PMC102730.

26. Markowski MC, Bowen C, Gelmann EP. 2008; Inflammatory cytokines induce phosphorylation and ubiquitination of prostate suppressor protein NKX3.1. Cancer Res. 68:6896–6901. DOI:
10.1158/0008-5472.CAN-08-0578. PMID:
18757402. PMCID:
PMC2586101.

27. Bethel CR, Faith D, Li X, Guan B, Hicks JL, Lan F, Jenkins RB, Bieberich CJ, De Marzo AM. 2006; Decreased NKX3.1 protein expression in focal prostatic atrophy, prostatic intraepithelial neoplasia, and adenocarcinoma: association with gleason score and chromosome 8p deletion. Cancer Res. 66:10683–10690. DOI:
10.1158/0008-5472.CAN-06-0963. PMID:
17108105.

28. Gelmann EP, Steadman DJ, Ma J, Ahronovitz N, Voeller HJ, Swope S, Abbaszadegan M, Brown KM, Strand K, Hayes RB, Stampfer MJ. 2002; Occurrence of NKX3.1 C154T polymorphism in men with and without prostate cancer and studies of its effect on protein function. Cancer Res. 62:2654–2659. PMID:
11980664.
29. Guan B, Pungaliya P, Li X, Uquillas C, Mutton LN, Rubin EH, Bieberich CJ. 2008; Ubiquitination by TOPORS regulates the prostate tumor suppressor NKX3.1. J Biol Chem. 283:4834–4840. DOI:
10.1074/jbc.M708630200. PMID:
18077445.

30. Bowen C, Stuart A, Ju JH, Tuan J, Blonder J, Conrads TP, Veenstra TD, Gelmann EP. 2007; NKX3.1 homeodomain protein binds to topoisomerase I and enhances its activity. Cancer Res. 67:455–464. DOI:
10.1158/0008-5472.CAN-06-1591. PMID:
17234752.

31. Song LN, Bowen C, Gelmann EP. 2013; Structural and functional interactions of the prostate cancer suppressor protein NKX3.1 with topoisomerase I. Biochem J. 453:125–136. DOI:
10.1042/BJ20130012. PMID:
23557481. PMCID:
PMC7711313.

32. Padmanabhan A, Gosc EB, Bieberich CJ. 2013; Stabilization of the prostate-specific tumor suppressor NKX3.1 by the oncogenic protein kinase Pim-1 in prostate cancer cells. J Cell Biochem. 114:1050–1057. DOI:
10.1002/jcb.24444. PMID:
23129228.

33. Aumüller G, Seitz J. 1990; Protein secretion and secretory processes in male accessory sex glands. Int Rev Cytol. 121:127–231. DOI:
10.1016/S0074-7696(08)60660-9. PMID:
2190945.

34. Ayala AG, Ro JY, Babaian R, Troncoso P, Grignon DJ. 1989; The prostatic capsule: does it exist? Its importance in the staging and treatment of prostatic carcinoma. Am J Surg Pathol. 13:21–27. DOI:
10.1097/00000478-198901000-00003. PMID:
2909195.
36. Lowsley OS. 1912; The development of the human prostate gland with reference to the development of other structures at the neck of the urinary bladder. Am J Anat. 13:299–349. DOI:
10.1002/aja.1000130303.

38. Cunha GR, Donjacour AA, Cooke PS, Mee S, Bigsby RM, Higgins SJ, Sugimura Y. 1987; The endocrinology and developmental biology of the prostate. Endocr Rev. 8:338–362. DOI:
10.1210/edrv-8-3-338. PMID:
3308446.

39. Price D. 1963; Comparative aspects of development and structure in the prostate. Natl Cancer Inst Monogr. 12:1–27. PMID:
14072991.
41. Bhatia-Gaur R, Donjacour AA, Sciavolino PJ, Kim M, Desai N, Young P, Norton CR, Gridley T, Cardiff RD, Cunha GR, Abate-Shen C, Shen MM. 1999; Roles for Nkx3.1 in prostate development and cancer. Genes Dev. 13:966–977. DOI:
10.1101/gad.13.8.966. PMID:
10215624. PMCID:
PMC316645.

43. Dutta A, Le Magnen C, Mitrofanova A, Ouyang X, Califano A, Abate-Shen C. 2016; Identification of an NKX3.1-G9a-UTY transcriptional regulatory network that controls prostate differentiation. Science. 352:1576–1580. DOI:
10.1126/science.aad9512. PMID:
27339988. PMCID:
PMC5507586.

44. Huang L, Pu Y, Alam S, Birch L, Prins GS. 2005; The role of Fgf10 signaling in branching morphogenesis and gene expression of the rat prostate gland: lobe-specific suppression by neonatal estrogens. Dev Biol. 278:396–414. DOI:
10.1016/j.ydbio.2004.11.020. PMID:
15680359.

45. Simons BW, Hurley PJ, Huang Z, Ross AE, Miller R, Marchionni L, Berman DM, Schaeffer EM. 2012; Wnt signaling though beta-catenin is required for prostate lineage specification. Dev Biol. 371:246–255. DOI:
10.1016/j.ydbio.2012.08.016. PMID:
22960283. PMCID:
PMC3472417.

46. Francis JC, Thomsen MK, Taketo MM, Swain A. 2013; β-catenin is required for prostate development and cooperates with Pten loss to drive invasive carcinoma. PLoS Genet. 9:e1003180. DOI:
10.1371/journal.pgen.1003180. PMID:
23300485. PMCID:
PMC3536663.

47. Zhang TJ, Hoffman BG, Ruiz de Algara T, Helgason CD. 2006; SAGE reveals expression of Wnt signalling pathway members during mouse prostate development. Gene Expr Patterns. 6:310–324. DOI:
10.1016/j.modgep.2005.07.005. PMID:
16378759.

48. Schaeffer EM, Marchionni L, Huang Z, Simons B, Blackman A, Yu W, Parmigiani G, Berman DM. 2008; Androgen-induced programs for prostate epithelial growth and invasion arise in embryogenesis and are reactivated in cancer. Oncogene. 27:7180–7191. DOI:
10.1038/onc.2008.327. PMID:
18794802. PMCID:
PMC2676849.

49. Yu X, Wang Y, Jiang M, Bierie B, Roy-Burman P, Shen MM, Taketo MM, Wills M, Matusik RJ. 2009; Activation of beta-Catenin in mouse prostate causes HGPIN and continuous prostate growth after castration. Prostate. 69:249–262. DOI:
10.1002/pros.20877. PMID:
18991257. PMCID:
PMC4437562.

50. Mehta V, Schmitz CT, Keil KP, Joshi PS, Abler LL, Lin TM, Taketo MM, Sun X, Vezina CM. 2013; Beta-catenin (CTNNB1) induces Bmp expression in urogenital sinus epithelium and participates in prostatic bud initiation and patterning. Dev Biol. 376:125–135. DOI:
10.1016/j.ydbio.2013.01.034. PMID:
23396188. PMCID:
PMC3602957.

53. Wang X, Kruithof-de Julio M, Economides KD, Walker D, Yu H, Halili MV, Hu YP, Price SM, Abate-Shen C, Shen MM. 2009; A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 461:495–500. DOI:
10.1038/nature08361. PMID:
19741607. PMCID:
PMC2800362.

54. Bowen C, Bubendorf L, Voeller HJ, Slack R, Willi N, Sauter G, Gasser TC, Koivisto P, Lack EE, Kononen J, Kallioniemi OP, Gelmann EP. 2000; Loss of NKX3.1 expression in human prostate cancers correlates with tumor pro-gression. Cancer Res. 60:6111–6115. PMID:
11085535.
55. Shen MM, Abate-Shen C. 2010; Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 24:1967–2000. DOI:
10.1101/gad.1965810. PMID:
20844012. PMCID:
PMC2939361.

56. Decker J, Jain G, Kießling T, Sander P, Rid M, Barth TTF, Möller P, Cronauer MV, Marienfeld RB. 2016; Loss of the tumor suppressor NKX3.1 in prostate cancer cells is induced by prostatitis related mitogens. J Clin Exp Oncol. 5:2. DOI:
10.4172/2324-9110.1000160.

57. Le Magnen C, Virk RK, Dutta A, Kim JY, Panja S, Lopez-Bujanda ZA, Califano A, Drake CG, Mitrofanova A, Abate-Shen C. 2018; Cooperation of loss of
NKX3.1 and inflammation in prostate cancer initiation. Dis Model Mech. 11:dmm035139. DOI:
10.1242/dmm.035139. PMID:
30266798. PMCID:
PMC6262819.
58. Song H, Zhang B, Watson MA, Humphrey PA, Lim H, Milbrandt J. 2009; Loss of Nkx3.1 leads to the activation of discrete downstream target genes during prostate tumorigene-sis. Oncogene. 28:3307–3319. DOI:
10.1038/onc.2009.181. PMID:
19597465. PMCID:
PMC2746257.

59. Peng M, Deng J, Zhou S, Tao T, Su Q, Yang X, Yang X. 2019; The role of Clusterin in cancer metastasis. Cancer Manag Res. 11:2405–2414. DOI:
10.2147/CMAR.S196273. PMID:
31114318. PMCID:
PMC6497892.
60. Soloviev M, Esteves MP, Amiri F, Crompton MR, Rider CC. 2013; Elevated transcription of the gene QSOX1 encoding quiescin Q6 sulfhydryl oxidase 1 in breast cancer. PLoS One. 8:e57327. DOI:
10.1371/journal.pone.0057327. PMID:
23460839. PMCID:
PMC3583868.

61. Dong JT. 2001; Chromosomal deletions and tumor suppressor genes in prostate cancer. Cancer Metastasis Rev. 20:173–193. DOI:
10.1023/A:1015575125780. PMID:
12085961.

62. Emmert-Buck MR, Vocke CD, Pozzatti RO, Duray PH, Jennings SB, Florence CD, Zhuang Z, Bostwick DG, Liotta LA, Linehan WM. 1995; Allelic loss on chromosome 8p12-21 in microdissected prostatic intraepithelial neoplasia. Cancer Res. 55:2959–2962. PMID:
7606709.
63. Akamatsu S, Takata R, Ashikawa K, Hosono N, Kamatani N, Fujioka T, Ogawa O, Kubo M, Nakamura Y, Nakagawa H. 2010; A functional variant in NKX3.1 associated with prostate cancer susceptibility down-regulates NKX3.1 expression. Hum Mol Genet. 19:4265–4272. DOI:
10.1093/hmg/ddq350. PMID:
20716579.

65. Suzuki H, Freije D, Nusskern DR, Okami K, Cairns P, Sidransky D, Isaacs WB, Bova GS. 1998; Interfocal heterogeneity of PTEN/MMAC1 gene alterations in multiple metastatic prostate cancer tissues. Cancer Res. 58:204–209. PMID:
9443392.
66. Kim MJ, Cardiff RD, Desai N, Banach-Petrosky WA, Parsons R, Shen MM, Abate-Shen C. 2002; Cooperativity of Nkx3.1 and Pten loss of function in a mouse model of prostate carcinogenesis. Proc Natl Acad Sci U S A. 99:2884–2889. DOI:
10.1073/pnas.042688999. PMID:
11854455. PMCID:
PMC122442.

67. Lei Q, Jiao J, Xin L, Chang CJ, Wang S, Gao J, Gleave ME, Witte ON, Liu X, Wu H. 2006; NKX3.1 stabilizes p53, inhibits AKT activation, and blocks prostate cancer initiation caused by PTEN loss. Cancer Cell. 9:367–378. DOI:
10.1016/j.ccr.2006.03.031. PMID:
16697957.

69. Kos L, Chiang C, Mahon KA. 1998; Mediolateral patterning of somites: multiple axial signals, including Sonic hedgehog, regulate Nkx-3.1 expression. Mech Dev. 70:25–34. DOI:
10.1016/S0925-4773(97)00168-8. PMID:
9510022.

70. Tanaka M, Lyons GE, Izumo S. 1999; Expression of the Nkx3.1 homobox gene during pre and postnatal development. Mech Dev. 85:179–182. DOI:
10.1016/S0925-4773(99)00084-2. PMID:
10415359.

71. Schneider A, Brand T, Zweigerdt R, Arnold H. 2000; Targeted disruption of the Nkx3.1 gene in mice results in morphogenetic defects of minor salivary glands: parallels to glandular duct morphogenesis in prostate. Mech Dev. 95:163–174. DOI:
10.1016/S0925-4773(00)00355-5. PMID:
10906459.

72. Treier M, Gleiberman AS, O'Connell SM, Szeto DP, McMahon JA, McMahon AP, Rosenfeld MG. 1998; Multistep signaling requirements for pituitary organogenesis in vivo. Genes Dev. 12:1691–1704. DOI:
10.1101/gad.12.11.1691. PMID:
9620855. PMCID:
PMC316866.
73. Miyaguchi K, Uzawa N, Mogushi K, Takahashi K, Michikawa C, Nakata Y, Sumino J, Okada N, Mizushima H, Fukuoka Y, Tanaka H. 2012; Loss of NKX3-1 as a potential marker for an increased risk of occult lymph node metastasis and poor prognosis in oral squamous cell carcinoma. Int J Oncol. 40:1907–1914. DOI:
10.3892/ijo.2012.1373. PMID:
22344708.

74. Takada N, Nishida H, Oyama Y, Kusaba T, Kadowaki H, Arakane M, Wada J, Urabe S, Daa T. 2020; Immunohisto-chemical reactivity of prostate-specific markers for Salivary duct carcinoma. Pathobiology. 87:30–36. DOI:
10.1159/000504810. PMID:
31865345.

75. Patel MS, Bowen DK, Tassone NM, Gould AD, Kochan KS, Firmiss PR, Kukulka NA, Devine MY, Li B, Gong EM, Dettman RW. 2019; The homeodomain transcription factor Nkx3.1 modulates bladder outlet obstruction induced fibrosis in mice. Front Pediatr. 7:446. DOI:
10.3389/fped.2019.00446. PMID:
31781523. PMCID:
PMC6861332.

76. Qian D, Zheng W, Chen C, Jing G, Huang J. 2020; Roles of CCNB2 and
NKX3-1 in nasopharyngeal carcinoma. Cancer Biother Radiopharm. 35:208–213. DOI:
10.1089/cbr.2019.3016. PMID:
32202926.
78. English HF, Santen RJ, Isaacs JT. 1987; Response of glandular versus basal rat ventral prostatic epithelial cells to androgen withdrawal and replacement. Prostate. 11:229–242. DOI:
10.1002/pros.2990110304. PMID:
3684783.

79. Evans GS, Chandler JA. 1987; Cell proliferation studies in the rat prostate: II. The effects of castration and androgen-induced regeneration upon basal and secretory cell prolifera-tion. Prostate. 11:339–351. DOI:
10.1002/pros.2990110406. PMID:
3684785.

80. Sugimura Y, Cunha GR, Donjacour AA. 1986; Morphological and histological study of castration-induced degeneration and androgen-induced regeneration in the mouse prostate. Biol Reprod. 34:973–983. DOI:
10.1095/biolreprod34.5.973. PMID:
3730489.

81. Wang ZA, Shen MM. 2011; Revisiting the concept of cancer stem cells in prostate cancer. Oncogene. 30:1261–1271. DOI:
10.1038/onc.2010.530. PMID:
21119602.

82. Germann M, Wetterwald A, Guzmán-Ramirez N, van der Pluijm G, Culig Z, Cecchini MG, Williams ED, Thalmann GN. 2012; Stem-like cells with luminal progenitor phenotype survive castration in human prostate cancer. Stem Cells. 30:1076–1086. DOI:
10.1002/stem.1087. PMID:
22438320.

83. Clark AK, Taubenberger AV, Taylor RA, Niranjan B, Chea ZY, Zotenko E, Sieh S, Pedersen JS, Norden S, Frydenberg M, Grummet JP, Pook DW, Stirzaker C, Clark SJ, Lawrence MG, Ellem SJ, Hutmacher DW, Risbridger GP. Australian Prostate Cancer BioResource. 2013; A bioengineered microenvironment to quantitatively measure the tumorigenic properties of cancer-associated fibroblasts in human prostate cancer. Biomaterials. 34:4777–4785. DOI:
10.1016/j.biomaterials.2013.03.005. PMID:
23562048.

84. Federer-Gsponer JR, Müller DC, Zellweger T, Eggimann M, Marston K, Ruiz C, Seifert HH, Rentsch CA, Bubendorf L, Le Magnen C. 2020; Patterns of stemness-associated markers in the development of castration-resistant prostate cancer. Prostate. 80:1108–1117. DOI:
10.1002/pros.24039. PMID:
32628318.

85. Adisetiyo H, Liang M, Liao CP, Jeong JH, Cohen MB, Roy-Burman P, Frenkel B. 2014; Dependence of castration-resistant prostate cancer (CRPC) stem cells on CRPC-associated fibroblasts. J Cell Physiol. 229:1170–1176. DOI:
10.1002/jcp.24546. PMID:
24752784. PMCID:
PMC5774665.

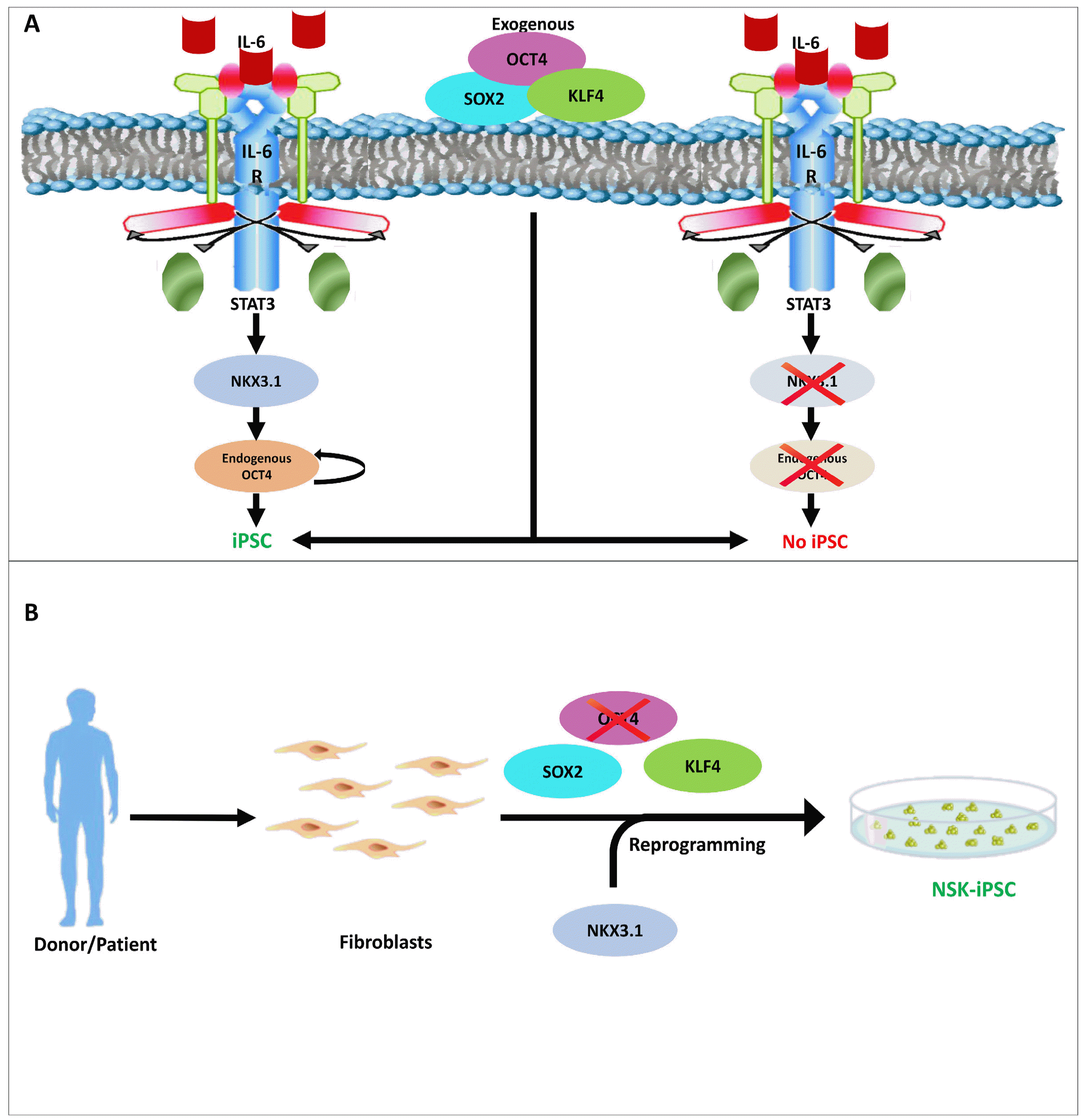
86. Mai T, Markov GJ, Brady JJ, Palla A, Zeng H, Sebastiano V, Blau HM. 2018; NKX3-1 is required for induced pluripotent stem cell reprogramming and can replace OCT4 in mouse and human iPSC induction. Nat Cell Biol. 20:900–908. DOI:
10.1038/s41556-018-0136-x. PMID:
30013107. PMCID:
PMC6101038.

87. Takahashi K, Yamanaka S. 2006; Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 126:663–676. DOI:
10.1016/j.cell.2006.07.024. PMID:
16904174.

88. Takahashi K, Yamanaka S. 2016; A decade of transcription factor-mediated reprogramming to pluripotency. Nat Rev Mol Cell Biol. 17:183–193. DOI:
10.1038/nrm.2016.8. PMID:
26883003.

89. Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A, Jaenisch R. 2012; Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 150:1209–1222. DOI:
10.1016/j.cell.2012.08.023. PMID:
22980981. PMCID:
PMC3457656.

90. Menendez S, Camus S, Herreria A, Paramonov I, Morera LB, Collado M, Pekarik V, Maceda I, Edel M, Consiglio A, Sanchez A, Li H, Serrano M, Belmonte JC. 2012; Increased dosage of tumor suppressors limits the tumorigenicity of iPS cells without affecting their pluripotency. Aging Cell. 11:41–50. DOI:
10.1111/j.1474-9726.2011.00754.x. PMID:
21981310.

91. Velychko S, Adachi K, Kim KP, Hou Y, MacCarthy CM, Wu G, Schöler HR. 2019; Excluding Oct4 from Yamanaka cocktail unleashes the developmental potential of iPSCs. Cell Stem Cell. 25:737–753.e4. DOI:
10.1016/j.stem.2019.10.002. PMID:
31708402. PMCID:
PMC6900749.

92. Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, Qin B, Xu J, Li W, Yang J, Gan Y, Qin D, Feng S, Song H, Yang D, Zhang B, Zeng L, Lai L, Esteban MA, Pei D. 2010; A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 7:51–63. DOI:
10.1016/j.stem.2010.04.014. PMID:
20621050.

93. Malik V, Glaser LV, Zimmer D, Velychko S, Weng M, Holzner M, Arend M, Chen Y, Srivastava Y, Veerapandian V, Shah Z, Esteban MA, Wang H, Chen J, Schöler HR, Hutchins AP, Meijsing SH, Pott S, Jauch R. 2019; Pluripotency reprogramming by competent and incompetent POU factors uncovers temporal dependency for Oct4 and Sox2. Nat Commun. 10:3477. DOI:
10.1038/s41467-019-11054-7. PMID:
31375664. PMCID:
PMC6677745.

94. Soufi A, Donahue G, Zaret KS. 2012; Facilitators and impediments of the pluripotency reprogramming factors' initial engagement with the genome. Cell. 151:994–1004. DOI:
10.1016/j.cell.2012.09.045. PMID:
23159369. PMCID:
PMC3508134.

96. Kuan II, Liang KH, Wang YP, Kuo TW, Meir YJ, Wu SC, Yang SC, Lu J, Wu HC. 2017; EpEX/EpCAM and Oct4 or Klf4 alone are sufficient to generate induced pluripotent stem cells through STAT3 and HIF2α. Sci Rep. 7:41852. DOI:
10.1038/srep41852. PMID:
28157205. PMCID:
PMC5291097.

98. Wenzel PL, Wu L, de Bruin A, Chong JL, Chen WY, Dureska G, Sites E, Pan T, Sharma A, Huang K, Ridgway R, Mosaliganti K, Sharp R, Machiraju R, Saltz J, Yamamoto H, Cross JC, Robinson ML, Leone G. 2007; Rb is critical in a mammalian tissue stem cell population. Genes Dev. 21:85–97. DOI:
10.1101/gad.1485307. PMID:
17210791. PMCID:
PMC1759903.

99. Mosteiro L, Pantoja C, de Martino A, Serrano M. 2018; Senescence promotes in vivo reprogramming through p16INK4a and IL-6. Aging Cell. 17:e12711. DOI:
10.1111/acel.12711. PMID:
29280266. PMCID:
PMC5847859.
100. Laurent LC, Ulitsky I, Slavin I, Tran H, Schork A, Morey R, Lynch C, Harness JV, Lee S, Barrero MJ, Ku S, Martynova M, Semechkin R, Galat V, Gottesfeld J, Izpisua Belmonte JC, Murry C, Keirstead HS, Park HS, Schmidt U, Laslett AL, Muller FJ, Nievergelt CM, Shamir R, Loring JF. 2011; Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell. 8:106–118. DOI:
10.1016/j.stem.2010.12.003. PMID:
21211785. PMCID:
PMC3043464.

101. Brady JJ, Li M, Suthram S, Jiang H, Wong WH, Blau HM. 2013; Early role for IL-6 signalling during generation of induced pluripotent stem cells revealed by heterokaryon RNA-Seq. Nat Cell Biol. 15:1244–1252. DOI:
10.1038/ncb2835. PMID:
23995732. PMCID:
PMC4100556.

102. Kim MJ, Bhatia-Gaur R, Banach-Petrosky WA, Desai N, Wang Y, Hayward SW, Cunha GR, Cardiff RD, Shen MM, Abate-Shen C. 2002; Nkx3.1 mutant mice recapitulate early stages of prostate carcinogenesis. Cancer Res. 62:2999–3004. PMID:
12036903.
103. Jiang AL, Hu XY, Zhang PJ, He ML, Kong F, Liu ZF, Yuan HQ, Zhang JY. 2005; Up-regulation of NKX3.1 expression and inhibition of LNCaP cell proliferation induced by an inhibitory element decoy. Acta Biochim Biophys Sin (Shanghai). 37:335–340. DOI:
10.1111/j.1745-7270.2005.00047.x. PMID:
15880262.

104. Zhang P, Liu W, Zhang J, Guan H, Chen W, Cui X, Liu Q, Jiang A. 2010; Gene expression profiles in the PC-3 human prostate cancer cells induced by NKX3.1. Mol Biol Rep. 37:1505–1512. DOI:
10.1007/s11033-009-9549-8. PMID:
19462257.

105. Song LN, Silva J, Koller A, Rosenthal A, Chen EI, Gelmann EP. 2015; The tumor suppressor NKX3.1 is targeted for degradation by DYRK1B kinase. Mol Cancer Res. 13:913–922. DOI:
10.1158/1541-7786.MCR-14-0680. PMID:
25777618. PMCID:
PMC4511920.

106. Kim JB, Greber B, Araúzo-Bravo MJ, Meyer J, Park KI, Zaehres H, Schöler HR. 2009; Direct reprogramming of human neural stem cells by OCT4. Nature. 461:649–643. DOI:
10.1038/nature08436. PMID:
19718018.

107. Kim JB, Sebastiano V, Wu G, Araúzo-Bravo MJ, Sasse P, Gentile L, Ko K, Ruau D, Ehrich M, van den Boom D, Meyer J, Hübner K, Bernemann C, Ortmeier C, Zenke M, Fleischmann BK, Zaehres H, Schöler HR. 2009; Oct4-induced pluripotency in adult neural stem cells. Cell. 136:411–419. DOI:
10.1016/j.cell.2009.01.023. PMID:
19203577.

108. Tsai SY, Bouwman BA, Ang YS, Kim SJ, Lee DF, Lemischka IR, Rendl M. 2011; Single transcription factor reprogramming of hair follicle dermal papilla cells to induced pluripotent stem cells. Stem Cells. 29:964–971. DOI:
10.1002/stem.649. PMID:
21563278.

109. Wu T, Wang H, He J, Kang L, Jiang Y, Liu J, Zhang Y, Kou Z, Liu L, Zhang X, Gao S. 2011; Reprogramming of trophoblast stem cells into pluripotent stem cells by Oct4. Stem Cells. 29:755–763. DOI:
10.1002/stem.617. PMID:
21305674.

110. Gao Y, Chen J, Li K, Wu T, Huang B, Liu W, Kou X, Zhang Y, Huang H, Jiang Y, Yao C, Liu X, Lu Z, Xu Z, Kang L, Chen J, Wang H, Cai T, Gao S. 2013; Replacement of Oct4 by Tet1 during iPSC induction reveals an important role of DNA methylation and hydroxymethylation in repro-gramming. Cell Stem Cell. 12:453–469. DOI:
10.1016/j.stem.2013.02.005. PMID:
23499384.

111. Heng JC, Feng B, Han J, Jiang J, Kraus P, Ng JH, Orlov YL, Huss M, Yang L, Lufkin T, Lim B, Ng HH. 2010; The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell. 6:167–174. DOI:
10.1016/j.stem.2009.12.009. PMID:
20096661.

112. Shi Y, Do JT, Desponts C, Hahm HS, Schöler HR, Ding S. 2008; A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell. 2:525–528. DOI:
10.1016/j.stem.2008.05.011. PMID:
18522845.





 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download