Abstract
The 2019-novel coronavirus (SARS-CoV-2) pneumonia epidemic is a thorny public health problem faced by health officials and a major cause of concern for health professionals. However, the currently used immortalized cell lines and animal models, though easy to manipulate, can not thoroughly simulate real viral activity due to a lack of target cells, species isolation, and insufficient adequate tissues and organs for clinical research. Organoid that emerges as an effective model and time-saving approach can simulate the viral life cycle in vitro and explore a therapeutic target for antiviral drug development. The 3D tissue cultures contain patient-specific stem cells in vitro to mimic the complexity of real tissue within the 3D microstructure that has the same functionality as the tissue of interest. It avoids the problems such as the distortion of genetic markers and animal ethics of using 2D cultures for animal testing and can be employed in studies of specific-organ viral infections to fully understand the physiopathological mechanism of SARS-CoV-2 infection for vaccine research and development.
The current coronavirus disease 2019 (COVID-19) epidemic caused by SARS-CoV-2 is life-threatening for all populations. There have been 71,581,532 confirmed cases and 1,618,374 deaths worldwide through 16 December, 2020 (1). As a highly pathogenic virus, preexisting evidence shows that SARS-CoV-2 cell entry lies in its strong affinity for the human angiotensin-converting enzyme 2 (ACE2) receptor through its receptor-binding region and the binding of the spines protein S to ACE 2 to activate the cell surface protease transmembrane serine protease family member II (TMPRSS2). This activation triggers viral-cell membrane fusion, which allows SARS-CoV-2 cell entry and initiates infection, it mainly causes alveolar injury and acute respiratory failure in patients, as well as varying degrees of damages to the nerve, circulation, digestion, urinary, and immune systems, which tremendously increases the probability to progress to a critical condition in patients with underlying diseases (2).
An in-vitro model shows the advantages of tracking the route for viral infection, identifying affected organs, exploring a series of mechanisms of pathological damage, and aiding in the development of vaccines and other interventions for precaution, control, and treatment for the infection (3). Chu et al. (4) analyzed 25 cell lines, and found that only the human lung (Caco2) and small intestinal (Calu3) cells are vulnerable to the virus, and they also found that none of the cell lines cultured under laboratory standard conditions could compare with those living in patients. In terms of the degree of cell immortalization, cultured cell lines cannot thoroughly mimic the actual inner environment of the organ in which cells of interest lived. Besides, a lack of cell lines or even one representative cell line in some organs often leads to the failure of the establishment of an organ culture system. These issues bias the experimentally predicted results from the clinical manifestations of viral infections. However, an obvious limitation of an animal model is that the symptoms of animal models cannot correspond to those of human patients at the same stages of the viral life cycle; so virus-related results can be biased in an all-around way (5). At present, because of the advantages in accessibility and operation, murines are commonly representative animal models, which are widely employed in virus research. However, species variation brings about categorical, distributional, and structural differences in ACE2 expression between humans versus mice. The use of transgenic technology in mice made ACE2 overexpression limited in the ileum; after the SARS-CoV-2 infection, the virus was merely detectable in the lung and the intestine, which was inconsistent with multisystem damage in infected patients (6); there even exist distinct descriptions of mouse strains, and some studies reported that the coronavirus could not infect wide-type mice or rats (7). Since the inflammatory cytokine storm theory is being known to more people, mouse models are inappropriate for research on immune responses to the SARS-CoV-2 infection due to a lack of key inflammatory mediators, such as interleukin IL-32 and IL-37. Neither are other animals due to part or all of the problems, which become evident obstacles to the grasping of knowledge on this virus in an all-around way and subsequent vaccine development (8).
As the confirmed cases and deaths are climbing at an alarming pace, it is very urgent to develop a new model to accurately simulate the biological characteristics of the virus in a short period, without the mentioned shortcomings of cell and animal models. Organoids are a useful research tool to culture patient-specific stem cells to develop into 3D organs with organ-like properties. The organoid graft has a long-term survival in vivo, and it can be used for screening of new drug candidates, without problems of potential species-related differences and ethical difficulties (9). In this sense, organs abundant in ACE2-ex-pressing cells are the gateway for virus invasion. In the human body, in addition to the lung, ACE2 is expressed in the heart, kidney, small intestine, blood vessels, testis, and other tissues, as well as choroid, cornea, and hepatobiliary organoids. Hence organoid offers the most relevant preclinical models to effectively explore and accurately understand the pathophysiological characteristics of SARS-CoV-2 infection and pinpoint possible targets of drug action in a short period, as illustrated in Table 1. In this paper, we reviewed culture protocols and the efficacy of currently reported COVID-19 organoids to encourage more in-vitro and in-vivo studies of shedding light on the mechanism of SARS-CoV-2 infection (Fig. 1).
Some patients infected with SARS-CoV-2 present with central nervous symptoms such as headache, nausea, and emesis. Brain CT and MRI findings revealed multiple hemorrhagic lesions, and the viral RNAs were detectable in the cerebrospinal fluid (10). Also, SARS-CoV-2 brain infection was reported in mouse models. Of note, it is the central nervous system (CNS) infection rather than respiratory infection that leads to death, which provides a new direction to study the mechanism of the virus with damages to multi-organ function (11). The published evidence is insufficient to figure out the origin of the virus and the exact pathogenesis of the impairment of brain function after infection. Some studies used brain-like organoids containing the cell lines differentiated from human pluripotent stem cells (hPSC) to assess the susceptibility of brain cells to SARS-CoV-2 and further clarify the disease progression (12, 13). In the study by Jacob et al. (14, 15), after infection with mutant forms of SARS-CoV-2, cerebral organoids containing neuronal cells (astrocytes or microglia) and organoids that are representative of a single brain region, such as the cortex, hippocampus, hypothalamus, and midbrain, showed distinct affinities for the virus. And the virus aggregation was only found in the choroid plexus of hippocampal organoids. The choroid organoids have been established to understand cerebral dysfunction in SARS-CoV-2 infection, and transcriptome sequencing revealed that AchE2, TMPRSS2, and NRP1 were highly expressed in SARS-CoV-2-infected choroid plexus epithelial cells. This suggests that choroid plexus epithelial cells are probably the gateway for the entry of the SARS-CoV-2 virus into the CNS, which allows a cytoplasmic continuity between infected cells, that is, the syncytium, thus increasing the infection efficiency between adjacent cells (16). Besides, it was reported that cerebral organoids containing infected neurons in the replicative phase resulted in losses of taste and smell, delirium, stroke, and other neurological damages in patients. Although cerebral organoids imply the possible sites of SARS-CoV-2 entry in the brain, it is too early to conclude that the virus can invade the brain. Besides, cerebral organoid models are still in their infancy. Due to a lack of other mesenchymal components in the choroid organoid, the blood-brain barrier (BBB) has not been well-formed to block foreign bodies. Secondly, whether the high-level expression of ACE2 receptors or a lack of BBB contributes to the high concentration of the virus remains uncertain. Thirdly, the time of the virus infection of brain organoids between the studies by different teams were different, so this model requires modification for long-term observation in the future.
At present, brain organoids are used for the screening of Sofosbuvir and other candicate drugs (17). Obviously, this platform will be much more perfect for relative application after inducing of properly organized regional identities in brain organoids,in particular aimed at lucubrating SARS-CoV-2 infection.
As the SARS-CoV-2 virus mainly infects the respiratory tract by aerosol, wearing a mask and social distancing to block the transmission have been undertaken. However, other studies indicated that ACE2 and TMPRSS2 were expressed on the ocular surface, and animal experiments showed that SARS-CoV-2 was detected positive in the conjunctiva of infected rhesus monkeys (18). Therefore, using eye organoids to predict the infection site is needed. Besides, protections against eye infection is non-negli-gible. Makovoz et al. (19) established six types of eye organoids cultured from human PSCs and found that ACE2 and TMPRSS2 were highly expressed in cornea organoids. After one hour of infection of corneal organoids by SARS-CoV-2 in vitro, the viral replication was detectable. This suggests that the cornea can be considered as the entrance and proliferation site of virus infection. At the same time, TMPRSS11E was detected in corneal organoids, which showed a high degree of structural similarity with ACE2 and a high identity with TMPRSS2 in the domain and functional sites. This highlights comprehensive protective measures against SARS-CoV-2 infection, including eye infection, and indicates the use of TMPRSS11E as a potential infection site for the corresponding inhibitor treatment (20). Besides, the inflammation storm induced by a high NF-KB level in this corneal organoid model suggests that whether the inflammation storm is caused by SARS-CoV-2 eye infection or spreading the virus to the eye via respiratory or systemic infections has not been fully understood, which needs further exploration.
Ocular organoids, it is warranted to better clarify how infection in the eye may be helpful to know the pathway of transmission into other regions of the body.
Clinically, 20%∼30% of infected patients showed severe damages to cardiac function, such as myocardial damage or even acute myocardial infarction; their MRI showed that 54% of these patients had myocardial edema (21). Given that the systemic circulation is closely related to the pulmonary circulation, ACE2 receptors are highly expressed in the heart. Hence, except for acute injuries to the respiratory system, after the infection of the host by SARS-CoV-2, tissues are probably damaged, triggering inflammatory cytokine storm and the resultant vasoconstriction and metabolic disturbance in myocardial cells due to anoxia. Both jointly lead to irreversible heart injuries. To develop therapeutic regimens, heart organoids, as a platform for drug screening, are conducive to the implementation of multiple drug experiments in batch. Mills et al. (22) identified that the BRD4 (bromodomain protein 4)-fibrosis/iNOS axis was a pivotal intracellular mediator of severe cardiac functional damages in humans or even death induced by cytokine storm using heart organoids, proteomic analysis, and viral sequencing. Therefore, the axis can be considered to be a INCB054329, and this agent is expected to be a candidate to reduce damage to cardiac function in COVID-19 patients (23). However, another problem is that symptoms of arrhythmia and myocardial infarction cannot be mimicked by heart organoids; exempt from elimination reaction as a result of the absence of metabolic organs, drug doses required for heart organoids are significantly higher than the corresponding blood concentrations in humans. All these require more discussions about the translation of appropriate doses of candidate agents into clinical application.
Generate a more complete heart organoids, from the cellular composition, functional architecture to the reproducibility with the 3D bioprinting technology, making the organoids more advanced to simulate the changes associated with infection in vivo (24).
After infection with the virus, 80% of patients developed clinical manifestations such as lung injury and acute respiratory failure, which are related to the high expressions of ACE2 and TMPRSS2 in the airway epithelium (25). However, cells within the respiratory epithelium derived from mouse models did not express ACE2; thus, adequate animal models should be carefully selected for the studies of the interaction between the lung and SARS-CoV-2 virus. Pulmonary organoid cultures provide a functional research model in vitro for similar studies of respiratory diseases, which have been used for the research of respiratory syncytial, avian influenza, and parainfluenza viruses (such as H1N1, H7N9, H3N2) (26, 27). Han et al. (28, 29) developed a bronchial epithelial cell culture model (or human bronchial organoid) and identified that its highly expressed ACE2 and TMPRSS2 were associated with cell phenotypes. This model may offers advantages in accurately assessing human lung cells infected with SARS-CoV-2. During close monitoring for this model, they found that a large number of SARS-CoV-2 viruses were spread to the culture medium after the bronchi organoid was infected, making it possible to observe the process of the virus spreading from the replicative to secretive phase in vitro. They cultured lung organoids to screen a variety of drug candidates using a 384-well plate and found that imatinib, mycophenolic acid, quinacrine hydrochloride, and chloroquine effectively blocked the ACE2 cleavage site, thereby inhibiting viral replication in vivo. In their study, camostat, an inhibitor of TMPRSS2, was used to assess its efficacy in inhibiting viral infection and disease progression; the results revealed approximately 50% inhibition by camostat, and strikingly, remarkably suppressed inflammation storm which had been initiated by the activation of the IFN-1 after SARS-CoV-2 infection, thus avoiding the aggravation of extensive damages to the respiratory system (30). Interestingly, nowadays androgen signaling as a vital modulator of ACE2 levels through target analysis of hit compounds, and antiandrogenic drugs, such as finasteride, dutasteride, can downregulate the expression of ACE2 in lung organoids, therefore, reducing the susceptibility of men to SARS-CoV-19 and enriching the antiviral drugs candidate category (31). However, as the sources of stem cells reported are differed from each other, the susceptibility of lung organoids to the SARS-CoV-2 virus, the expression of virus-binding receptors, and the damage to different lung organoids after infection need to be further studied (32). Besides, no consensus on protocols for lung organoid cultures has been reached among different laboratories, and the longest cell culture time was even as long as 50 days, which may bias the experimental results as a whole.
The lung organoids are mainly accustomed for screening the FDA approved antivirual drug, with the tissue engineering and precision medicine popular, this organoid model should realize various exploration of pulmonary pathogens, especially during the outbreak period of SARS-CoV-2.
In addition to the most common respiratory virus infections, the number of clinical cases associated with the gastrointestinal tract is also on the rise. SARS-CoV-2 RNA was frequently detected in the feces of patients, some of whom with a negative nasopharyngeal test even showed positive anal swab. Current evidence reveals that mice can be infected with the virus by gavage, featuring symptoms of pulmonary infection (33). Along with the known evidence that ACE2 is a significant regulator of intestinal inflammation and is highly expressed in the small intestine, all this implies the gastrointestinal infection or potential fecal-oral transmission. The gut organoid that fueled a research boom of norovirus, Zika virus, avian H7N9, and swine H1N1 influenza is currently used for COVID-19 (34). Given the 96% homology between SARS-CoV-2 and isolated coronavirus Bat-SL-CoVZC45 and Bat-SL-CoVZXC21 from bats, Zhou et al. (35), for the first time, developed both human and bat intestinal organoid cultures and monitored ACE2, TMPRSS2, and cathepsin L levels at different time points after infection with SARS-CoV-2. The results showed that ACE2, TMPRSS2, and cathepsin L were highly expressed in intestinal organoids of humans and bats. Both human and bat intestinal organoid cultures presented a high copy number and a similar proliferation rate at the same time point. This study serves as a warning about solicitous protections for gastroscopists against both potential fecal-oral transmission and aerosol transmission when examining infected patients (36). Besides, expressions of some inflammatory mediators (IFNL2 and IFNL3) were up-regulated in human intestinal organoid cultures after SARS-CoV-2 infection, and whether the up-regulation of these factors in response to the host mediates the occurrence of corresponding clinical symptoms such as gastrointestinal syndromes should be confirmed (37).
Gastrointestinal organoids are mainly used for the identification and validation of virus infection of the digestive tract pathways and mechanisms, however, patients have two major mechanisms of antiviral ex vitro, intestinal flora and lymphatic system, namely defense barrier, regulating neutralization and destroying infection with outstanding contribution, further improve the structure of gastrointestinal organoids to be suitable for COVID-19 (38, 39).
Clinically, more than 50% of COVID-19 patients exhibit elevated serum levels of ALT, AST, albumin, and bilirubin, which indicate an underlying relationship between infection and liver injury. Relevant studies reported that ACE2 is only expressed in bile duct epithelial cells among digestive glands (40). Liver and bile duct organoid cultures are effective and time-saving methods to confirm the hypothesis that SARS-CoV-2 infection of the liver and biliary duct might indirectly cause liver injury. Yang et al. (41) established hepatobiliary organoids from bile duct epithelial cells and found the high expressions of ACE2 and TMPRSS2 in the organoid. After 24 hours of viral infection, qPCR analysis revealed high levels of viral replication, as well as up-regulated expression levels of apoptosis genes and down-regulated levels of the bile acid transport gene ACTB, ultimately leading to programmed death of hepatobiliary cells. Extensive apoptosis of hepatobiliary cells will break the bile duct epithelial barrier, resulting in bile acid accumulation and a series of clinical symptoms (42). Although significant up-regulation of chemokines in bile duct organoid cultures were observed, indicating the activation of inflammatory pathways in response to the viral infection, whether liver damage was directly caused by SARS-CoV-2 infection, inflammation storm, side effects from anti-SARS-CoV-2 drugs, hepatobiliary complications due to bile duct epithelial cell damage, or combined factors remains unknown. Moreover, this hepatobiliary organoid from human PSCs did not consist of important components of the immune system such as vascular endothelial cells and Kuffer cells, which cannot completely mimic human liver tissues rich in blood supply and the sites of the specific immune response following SARS-CoV-2 infection. Therefore, a modified hepatobiliary organoid with sufficient liver functions is required.
Hepatobiliary organoids provide insights into SARS-CoV-2 cell tropism, viral replication kinetics and the associated cellular responses after viral entry. However, considering that the cellular complexity of native tissues, for instance specific immune cell subsets, studys the interactions between human hepatobiliary epithelial cells and immune cells in the presence and absence of pathogens of COVID-19, Liver duct organoids will be commited into that to improve the states (41).
COVID-19 patients frequently reported hematuria, proteinuria, and other classic symptoms of acute kidney injury (43). Consistently, relevant studies have confirmed that ACE2 is widely expressed in the kidney, particularly in vascular endothelial cells within the kidney, this indicates that the kidney is vulnerable to the SARS-CoV-2 attack (44). Xia et al. (45) established organoid cultures of human proximal tubular epithelial cells combined with gene-editing techniques. They found that ACE2 expression was two times higher in 3D cultured cells than that in 2D cultured cells, suggesting that 3D culture conditions might be more pathologically relevant than 2D culture conditions. Monteil et al. (46) developed a kidney organoid model with high ACE2 expressions, which was highly consistent with the renal tissue structure within two weeks after six days of SARS-CoV-2 infection, the level of SARS-CoV-2 RNA in the supernatant of the renal organoid markedly increased, indicating that the kidney was vulnerable to the virus invasion. To illustrate whether SARS-CoV-2 invades renal cells via ACE2, they added a trial drug human recombinant soluble ACE2 (hrsACE2) to competitively binding to the virus rather than host cells (47). As a result, the SARS-CoV-2 load decreased by about 1000∼5000 times in a dose-dependent manner compared with the blank control group. The question is that current renal organoid cultures only mimic the renal functions of human fetuses in the second trimester, other than the infected kidney of adults. This encourages modified individual renal organoids targeting susceptible populations to understand renal infection with SARS-CoV-2 and the resultant renal dysfunction more accurately. As renal organoid models have been used for the validation of the efficacy of various trial drugs, whether the inhibition of virus load by hrsACE2 gives way to multiple viral invasions and whether it may induce antibody-dependent enhancement are two great concerns, which are critical to the vaccine development (48).
Kidney organoids containing relatively abundant ACE2 may be another viscera of virus attacks, in addition, antiviral agents cause tissue organs injuries to some degree, so the novel model lays the foundation for clinical combination experiment as a object for screening drug replacing cell lines and animals.
Infection of a 3D organoid model with viruses aids in the study of cell/tissue dynamics in an organoid. Besides, differences in drug diffusion and drug delivery to various tissues can be minimized by enhanced cell-to-cell contact within the model. Studies of the life cycle of SARS-CoV-2 and the efficacy of prevention and treatment drugs can be more objective by using 3D organoid models, with abundantly expressed ACE2 and TMPRSS2 in the models from human cells of interest. Theoretically, 3D organoid models perfectly mimic the complexity of a multi-cell environment, tissue structures, and functions of target organs to explore SARS-CoV-2 infection and drug screening more accurately (49). It is worth mentioning that SARS-CoV-2 often leads to type 1 diabetes in the clinic. Takebe et al. (50) developed pancreatic organoids from co-cultured PSCs, umbilical cord-derived endothelial cells, and mesenchymal stem cells, and transplanted the organoids into diabetic mice. They found that blood vessels in the host quickly grew into the organoid grafts, forming functional structures of the pancreas. Finally, the blood glucose levels and the weight of these diabetic mice returned to normal levels. This model renews hope for curing type I diabetes induced by SARS-CoV-2 infection. Currently, the failure rate of drug development during the preclinical testing period is 96.4% (51); 2D cell line cultures, on the other hand, as experimental models for drug testing, have significant differences in substrate mechanics compared with an internal microenvironment and genetic information, as well as flaws such as an individual mismatch. At the same time, a large amount of use of animals merely provides insufficient efforts in the progress of speedy approval of drugs for sale. So organoids, as an emerging model for virus research, have higher plasticity.
Considering that mucous membrane, membrane lining body cavities, and canals of the respiratory, digestive, and urogenital systems, as the first line of defense, are exposed to the outside world, protections against viral penetration into the first line of immune defense is a high priority (52). A problem of current organoid models for COVID-19 research is a lack of the involvement of innate immune cells. Therefore, a modified organoid model integrated with epithelium and immune cells is required to simulate natural immunity to SARS-CoV-2, and the dynamic process of the immune system coinciding with the disease progresses. A more convincing model will aid in future clinical trials to ascertain the efficacy and safety of treatment plans. Of note, current organoid models are devoid of components of multiple organs, which have not yet reflected organ-to-organ immunological communication (53). As COVID-19 is a multi-organ metabolic disease, studies of pathogenetic and antiviral mechanisms after infection require multi-purpose organoid models to responses and evaluate the efficacy and safety before clinical trials. Besides, notwithstanding cost-consuming and ethic and strain issues, animal models have obvious superiority over organoids in simulating infection signs after the viral invasion and establishing models of underlying diseases to explore possibly aggregated symptoms or even death after infection. At the same time, current technologies cannot yet simulate endocrine and nervous regulations in organoids and interactions between organoids and intestinal microecological conditions. Animal models can not only compensate these shortcomings of organoids but provide first-hand data of possible neuropsychiatric symptoms after virus infection as animals have behavioral and cognitive competences, which is a problem of organoids, as an emerging model for virus research, awaiting resolutions in comparison with animal models (8).
Organoids were ranked as one of Science’s Top 10 Breakthroughs of 2013. So far, a variety of organoids with main physiological structures and partial functions have been successfully cultivated, such as cerebral, lung, and gastrointestinal models. Its combination with gene-editing technologies such as CRISPR/Cas9 and biomaterials such as Nichoid, it is a three-dimensional scaffold based on the precise geometry of pores (54). The cells grown on the scaffold are subjected to the combined action of external forces such as mechanical load of fluid shear force and osmotic force, which mainly affects the cytoskeleton to induce gene rearrangement and thus lead to changes in the entire transcriptome (55). Therefore, organoids will be improved with the assistance of Nichoid and organoids-on-a-chip platforms that fine-regulate the physical and chemical parameters of organoid (53). As a carrier to study the high-throughput drug screening and the establishment of organoid biobanks and other techniques has achieved fruitful results (24). Therefore, the application of organoids to COVID-19 research is conducive to mimic processes from mastering the infection mechanism to developing therapeutic measures. Advances on continuously modified organoid models integrated with multiple cell components alongside other state-of-the-art technologies to overcome current shortcomings are helpful to provide a more reliable model for the in-depth study of the COVID-19 epidemic and accelerate the pace of the treatment.
References
1. World Health Organization. 2020. Dec. 16. Coronavirus disease (COVID-19) pandemic [Internet]. World Health Organization;Geneva: Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019. cited 2020 Dec 16.
2. Renu K, Prasanna PL, Valsala Gopalakrishnan A. 2020; Corona-viruses pathogenesis, comorbidities and multi-organ damage-a review. Life Sci. 255:117839. DOI: 10.1016/j.lfs.2020.117839. PMID: 32450165. PMCID: PMC7243768.
3. Sun SH, Chen Q, Gu HJ, Yang G, Wang YX, Huang XY, Liu SS, Zhang NN, Li XF, Xiong R, Guo Y, Deng YQ, Huang WJ, Liu Q, Liu QM, Shen YL, Zhou Y, Yang X, Zhao TY, Fan CF, Zhou YS, Qin CF, Wang YC. 2020; A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe. 28:124–133.e4. DOI: 10.1016/j.chom.2020.05.020. PMID: 32485164. PMCID: PMC7250783.

4. Chu H, Chan JF, Yuen TT, Shuai H, Yuan S, Wang Y, Hu B, Yip CC, Tsang JO, Huang X, Chai Y, Yang D, Hou Y, Chik KK, Zhang X, Fung AY, Tsoi HW, Cai JP, Chan WM, Ip JD, Chu AW, Zhou J, Lung DC, Kok KH, To KK, Tsang OT, Chan KH, Yuen KY. 2020; Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe. 1:e14–e23. DOI: 10.1016/S2666-5247(20)30004-5. PMID: 32835326. PMCID: PMC7173822.

5. Shi J, Wen Z, Zhong G, Yang H, Wang C, Huang B, Liu R, He X, Shuai L, Sun Z, Zhao Y, Liu P, Liang L, Cui P, Wang J, Zhang X, Guan Y, Tan W, Wu G, Chen H, Bu Z. 2020; Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 368:1016–1020. DOI: 10.1126/science.abb7015. PMID: 32269068. PMCID: PMC7164390.

6. Cohen J. 2020; From mice to monkeys, animals studied for coronavirus answers. Science. 368:221–222. DOI: 10.1126/science.368.6488.221. PMID: 32299928.

7. Hogan RJ, Gao G, Rowe T, Bell P, Flieder D, Paragas J, Kobinger GP, Wivel NA, Crystal RG, Boyer J, Feldmann H, Voss TG, Wilson JM. 2004; Resolution of primary severe acute respiratory syndrome-associated coronavirus infection requires Stat1. J Virol. 78:11416–11421. DOI: 10.1128/JVI.78.20.11416-11421.2004. PMID: 15452265. PMCID: PMC521834.

8. Johansen MD, Irving A, Montagutelli X, Tate MD, Rudloff I, Nold MF, Hansbro NG, Kim RY, Donovan C, Liu G, Faiz A, Short KR, Lyons JG, McCaughan GW, Gorrell MD, Cole A, Moreno C, Couteur D, Hesselson D, Triccas J, Neely GG, Gamble JR, Simpson SJ, Saunders BM, Oliver BG, Britton WJ, Wark PA, Nold-Petry CA, Hansbro PM. 2020; Animal and translational models of SARS-CoV-2 infection and COVID-19. Mucosal Immunol. 13:877–891. DOI: 10.1038/s41385-020-00340-z. PMID: 32820248. PMCID: PMC7439637.

9. Lancaster MA, Huch M. 2019; Disease modelling in human organoids. Dis Model Mech. 12:dmm039347. DOI: 10.1242/dmm.039347. PMID: 31383635. PMCID: PMC6679380.

10. Poyiadji N, Shahin G, Noujaim D, Stone M, Patel S, Griffith B. 2020; COVID-19-associated acute hemorrhagic necrotizing encephalopathy: imaging features. Radiology. 296:E119–E120. DOI: 10.1148/radiol.2020201187. PMID: 32228363. PMCID: PMC7233386.
11. Gu J, Gong E, Zhang B, Zheng J, Gao Z, Zhong Y, Zou W, Zhan J, Wang S, Xie Z, Zhuang H, Wu B, Zhong H, Shao H, Fang W, Gao D, Pei F, Li X, He Z, Xu D, Shi X, Anderson VM, Leong AS. 2005; Multiple organ infection and the pathogenesis of SARS. J Exp Med. 202:415–424. DOI: 10.1084/jem.20050828. PMID: 16043521. PMCID: PMC2213088.

12. Pellegrini L, Bonfio C, Chadwick J, Begum F, Skehel M, Lancaster MA. 2020; Human CNS barrier-forming organoids with cerebrospinal fluid production. Science. 369:eaaz5626. DOI: 10.1126/science.aaz5626. PMID: 32527923. PMCID: PMC7116154.

13. Pellegrini L, Albecka A, Mallery DL, Kellner MJ, Paul D, Carter AP, James LC, Lancaster MA. 2020; SARS-CoV-2 infects the brain choroid plexus and disrupts the blood-CSF barrier in human brain organoids. Cell Stem Cell. 27:951–961.e5. DOI: 10.1016/j.stem.2020.10.001. PMID: 33113348. PMCID: PMC7553118.

14. Jacob F, Pather SR, Huang WK, Zhang F, Wong SZH, Zhou H, Cubitt B, Fan W, Chen CZ, Xu M, Pradhan M, Zhang DY, Zheng W, Bang AG, Song H, Carlos de la Torre J, Ming GL. 2020; Human pluripotent stem cell-derived neural cells and brain organoids reveal SARS-CoV-2 neurotropism predominates in choroid plexus epithelium. Cell Stem Cell. 27:937–950.e9. DOI: 10.1016/j.stem.2020.09.016. PMID: 33010822. PMCID: PMC7505550.

15. Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, Yao B, Hamersky GR, Jacob F, Zhong C, Yoon KJ, Jeang W, Lin L, Li Y, Thakor J, Berg DA, Zhang C, Kang E, Chickering M, Nauen D, Ho CY, Wen Z, Christian KM, Shi PY, Maher BJ, Wu H, Jin P, Tang H, Song H, Ming GL. 2016; Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 165:1238–1254. DOI: 10.1016/j.cell.2016.04.032. PMID: 27118425. PMCID: PMC4900885.

16. Zhang BZ, Chu H, Han S, Shuai H, Deng J, Hu YF, Gong HR, Lee AC, Zou Z, Yau T, Wu W, Hung IF, Chan JF, Yuen KY, Huang JD. 2020; SARS-CoV-2 infects human neural progenitor cells and brain organoids. Cell Res. 30:928–931. DOI: 10.1038/s41422-020-0390-x. PMID: 32753756. PMCID: PMC7399356.

17. Mesci P, Macia A, Saleh A, Martin-Sancho L, Yin X, Snethlage C, Avansini S, Chanda SK, Muotri A. 2020; Sofosbuvir protects human brain organoids against SARS-CoV-2. bioRxiv 125856 [Preprint]. Available from: https://doi.org/10.1101/2020.05.30.125856. cited 2020 Dec 16. DOI: 10.1101/2020.05.30.125856.

18. Aiello F, Gallo Afflitto G, Mancino R, Li JO, Cesareo M, Giannini C, Nucci C. 2020; Coronavirus disease 2019 (SARS-CoV-2) and colonization of ocular tissues and secretions: a systematic review. Eye (Lond). 34:1206–1211. DOI: 10.1038/s41433-020-0926-9. PMID: 32424327. PMCID: PMC7232062.

19. Makovoz B, Moeller R, Zebitz Eriksen A, tenOever BR, Blenkinsop TA. 2020; SARS-CoV-2 infection of ocular cells from human adult donor eyes and hESC-derived eye organoids. SSRN 3650574 [Preprint]. Available from: https://doi.org/10.2139/ssrn.3650574. cited 2020 Dec 16. DOI: 10.2139/ssrn.3650574. PMID: 32742243. PMCID: PMC7385483.

20. Lai THT, Tang EWH, Chau SKY, Fung KSC, Li KKW. 2020; Stepping up infection control measures in ophthalmology during the novel coronavirus outbreak: an experience from Hong Kong. Graefes Arch Clin Exp Ophthalmol. 258:1049–1055. DOI: 10.1007/s00417-020-04641-8. PMID: 32124000. PMCID: PMC7087729.

21. Huang L, Zhao P, Tang D, Zhu T, Han R, Zhan C, Liu W, Zeng H, Tao Q, Xia L. 2020; Cardiac involvement in patients recovered from COVID-2019 identified using magnetic resonance imaging. JACC Cardiovasc Imaging. 13:2330–2339. DOI: 10.1016/j.jcmg.2020.05.004. PMID: 32763118. PMCID: PMC7214335.

22. Mills RJ, Titmarsh DM, Koenig X, Parker BL, Ryall JG, Quaife-Ryan GA, Voges HK, Hodson MP, Ferguson C, Drowley L, Plowright AT, Needham EJ, Wang QD, Gregorevic P, Xin M, Thomas WG, Parton RG, Nielsen LK, Launikonis BS, James DE, Elliott DA, Porrello ER, Hudson JE. 2017; Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc Natl Acad Sci U S A. 114:E8372–E8381. DOI: 10.1073/pnas.1707316114. PMID: 28916735. PMCID: PMC5635889.

23. Mills RJ, Humphrey SJ, Fortuna PRJ, Quaife-Ryan GA, Mehdiabadi NR, Devilée L, Voges HK, Reynolds LT, Krumeich S, Mathieson E, Griffen B, Titmarsh D, Porrello ER, Smyth MJ, Engwerda CR, MacDonald KPA, Bald T, James DE, Hudson JE. 2020; Discovery of drugs to treat cytokine storm-induced cardiac dysfunction using human cardiac organoids. bioRxiv 258574 [Preprint]. Available from: https://doi.org/10.1101/2020.08.23.258574. cited 2020 Dec 16. DOI: 10.1101/2020.08.23.258574.

24. Garreta E, Kamm RD, Chuva de Sousa Lopes SM, Lancaster MA, Weiss R, Trepat X, Hyun I, Montserrat N. 2020; Rethinking organoid technology through bioengineering. Nat Mater. [Epub ahead of print]. DOI: 10.1038/s41563-020-00804-4. PMID: 33199860.

25. Ackermann M, Verleden SE, Kuehnel M, Haverich A, Welte T, Laenger F, Vanstapel A, Werlein C, Stark H, Tzankov A, Li WW, Li VW, Mentzer SJ, Jonigk D. 2020; Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N Engl J Med. 383:120–128. DOI: 10.1056/NEJMoa2015432. PMID: 32437596. PMCID: PMC7412750.

26. Zhou J, Li C, Sachs N, Chiu MC, Wong BH, Chu H, Poon VK, Wang D, Zhao X, Wen L, Song W, Yuan S, Wong KK, Chan JF, To KK, Chen H, Clevers H, Yuen KY. 2018; Differentiated human airway organoids to assess infectivity of emerging influenza virus. Proc Natl Acad Sci U S A. 115:6822–6827. DOI: 10.1073/pnas.1806308115. PMID: 29891677. PMCID: PMC6042130.

27. Porotto M, Ferren M, Chen YW, Siu Y, Makhsous N, Rima B, Briese T, Greninger AL, Snoeck HW, Moscona A. 2019; Authentic modeling of human respiratory virus infection in human pluripotent stem cell-derived lung organoids. mBio. 10:e00723–19. DOI: 10.1128/mBio.00723-19. PMID: 31064833. PMCID: PMC6509192.

28. Han Y, Yang L, Duan X, Duan F, Nilsson-Payant BE, Yaron TM, Wang P, Tang X, Zhang T, Zhao Z, Bram Y, Redmond D, Houghton S, Nguyen D, Xu D, Wang X, Uhl S, Huang Y, Johnson JL, Xiang J, Wang H, Pan FC, Cantley LC, tenOever BR, Ho DD, Evans T, Schwartz RE, Chen HJ, Chen S. 2020; Identification of candidate COVID-19 therapeutics using hPSC-derived lung organoids. bioRxiv 079095 [Preprint]. Available from: https://doi.org/10.1101/2020.05.05.079095. cited 2020 Dec 16. DOI: 10.1101/2020.05.05.079095. PMID: 32511403. PMCID: PMC7263550.

29. Han Y, Duan X, Yang L, Nilsson-Payant BE, Wang P, Duan F, Tang X, Yaron TM, Zhang T, Uhl S, Bram Y, Richardson C, Zhu J, Zhao Z, Redmond D, Houghton S, Nguyen DT, Xu D, Wang X, Jessurun J, Borczuk A, Huang Y, Johnson JL, Liu Y, Xiang J, Wang H, Cantley LC, tenOever BR, Ho DD, Pan FC, Evans T, Chen HJ, Schwartz RE, Chen S. 2020; Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature. [Epub ahead of print]. DOI: 10.1038/s41586-020-2901-9. PMID: 33116299. PMCID: PMC8034380.

30. Elbadawi M, Efferth T. 2020; Organoids of human airways to study infectivity and cytopathy of SARS-CoV-2. Lancet Respir Med. 8:e55–e56. DOI: 10.1016/S2213-2600(20)30238-1. PMID: 32446313. PMCID: PMC7241998.

31. Samuel RM, Majd H, Richter MN, Ghazizadeh Z, Zekavat SM, Navickas A, Ramirez JT, Asgharian H, Simoneau CR, Bonser LR, Koh KD, Garcia-Knight M, Tassetto M, Sunshine S, Farahvashi S, Kalantari A, Liu W, Andino R, Zhao H, Natarajan P, Erle DJ, Ott M, Goodarzi H, Fattahi F. 2020; Androgen signaling regulates SARS-CoV-2 receptor levels and is associated with severe COVID-19 symptoms in men. Cell Stem Cell. 27:876–889.e12. DOI: 10.1016/j.stem.2020.11.009. PMID: 33232663. PMCID: PMC7670929.

32. Hui KPY, Ching RHH, Chan SKH, Nicholls JM, Sachs N, Clevers H, Peiris JSM, Chan MCW. 2018; Tropism, replication competence, and innate immune responses of influenza virus: an analysis of human airway organoids and ex-vivo bronchus cultures. Lancet Respir Med. 6:846–854. DOI: 10.1016/S2213-2600(18)30236-4. PMID: 30001996.

33. Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. 2020; Evidence for gastrointestinal infection of SARS-CoV-2. Gastroentero-logy. 158:1831–1833.e3. DOI: 10.1053/j.gastro.2020.02.055. PMID: 32142773. PMCID: PMC7130181.

34. Bertram S, Heurich A, Lavender H, Gierer S, Danisch S, Perin P, Lucas JM, Nelson PS, Pöhlmann S, Soilleux EJ. 2012; Influenza and SARS-coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PLoS One. 7:e35876. DOI: 10.1371/journal.pone.0035876. PMID: 22558251. PMCID: PMC3340400.

35. Zhou J, Li C, Liu X, Chiu MC, Zhao X, Wang D, Wei Y, Lee A, Zhang AJ, Chu H, Cai JP, Yip CC, Chan IH, Wong KK, Tsang OT, Chan KH, Chan JF, To KK, Chen H, Yuen KY. 2020; Infection of bat and human intestinal organoids by SARS-CoV-2. Nat Med. 26:1077–1083. DOI: 10.1038/s41591-020-0912-6. PMID: 32405028.

36. Dickson I. 2020; Organoids demonstrate gut infection by SARS-CoV-2. Nat Rev Gastroenterol Hepatol. 17:383. DOI: 10.1038/s41575-020-0317-5. PMID: 32427981. PMCID: PMC7235550.

37. Mallapaty S. 2020; Mini organs reveal how the coronavirus ravages the body. Nature. 583:15–16. DOI: 10.1038/d41586-020-01864-x. PMID: 32606457.

38. Lamers MM, Beumer J, van der Vaart J, Knoops K, Puschhof J, Breugem TI, Ravelli RBG, Paul van Schayck J, Mykytyn AZ, Duimel HQ, van Donselaar E, Riesebosch S, Kuijpers HJH, Schipper D, van de Wetering WJ, de Graaf M, Koopmans M, Cuppen E, Peters PJ, Haagmans BL, Clevers H. 2020; SARS-CoV-2 productively infects human gut enterocytes. Science. 369:50–54. DOI: 10.1126/science.abc1669. PMID: 32358202. PMCID: PMC7199907.

39. Krüger J, Groß R, Conzelmann C, Müller JA, Koepke L, Sparrer KMJ, Weil T, Schütz D, Seufferlein T, Barth TFE, Stenger S, Heller S, Münch J, Kleger A. 2020; Drug inhibition of SARS-CoV-2 replication in human pluripotent stem cell-derived intestinal organoids. Cell Mol Gastroenterol Hepatol. [Epub ahead of print]. DOI: 10.1101/2020.06.10.144816. PMID: 33186749. PMCID: PMC7655023.

40. Jothimani D, Venugopal R, Abedin MF, Kaliamoorthy I, Rela M. 2020; COVID-19 and the liver. J Hepatol. 73:1231–1240. DOI: 10.1016/j.jhep.2020.06.006. PMID: 32553666. PMCID: PMC7295524.

41. Yang L, Han Y, Nilsson-Payant BE, Gupta V, Wang P, Duan X, Tang X, Zhu J, Zhao Z, Jaffré F, Zhang T, Kim TW, Harschnitz O, Redmond D, Houghton S, Liu C, Naji A, Ciceri G, Guttikonda S, Bram Y, Nguyen DT, Cioffi M, Chandar V, Hoagland DA, Huang Y, Xiang J, Wang H, Lyden D, Borczuk A, Chen HJ, Studer L, Pan FC, Ho DD, tenOever BR, Evans T, Schwartz RE, Chen S. 2020; A human pluripotent stem cell-based platform to study SARS-CoV-2 tropism and model virus infection in human cells and organoids. Cell Stem Cell. 27:125–136.e7. DOI: 10.1016/j.stem.2020.06.015. PMID: 32579880. PMCID: PMC7303620.

42. Zhao B, Ni C, Gao R, Wang Y, Yang L, Wei J, Lv T, Liang J, Zhang Q, Xu W, Xie Y, Wang X, Yuan Z, Liang J, Zhang R, Lin X. 2020; Recapitulation of SARS-CoV-2 infection and cholangiocyte damage with human liver ductal organoids. Protein Cell. 11:771–775. DOI: 10.1007/s13238-020-00718-6. PMID: 32303993. PMCID: PMC7164704.

43. Qian JY, Wang B, Liu BC. 2020; Acute kidney injury in the 2019 novel coronavirus disease. Kidney Dis (Basel). 323:1–6. DOI: 10.1159/000509086. PMID: 32742978. PMCID: PMC7360511.

44. Shetty AA, Tawhari I, Safar-Boueri L, Seif N, Alahmadi A, Gargiulo R, Aggarwal V, Usman I, Kisselev S, Gharavi AG, Kanwar Y, Quaggin SE. 2021; COVID-19-associated glomerular disease. J Am Soc Nephrol. 32:33–40. DOI: 10.1681/ASN.2020060804. PMID: 33214201. PMCID: PMC7894674.

45. Xia S, Wu M, Chen S, Zhang T, Ye L, Liu J, Li H. 2020; Long term culture of human kidney proximal tubule epithelial cells maintains lineage functions and serves as an ex vivo model for coronavirus associated kidney injury. Virol Sin. 35:311–320. DOI: 10.1007/s12250-020-00253-y. PMID: 32602046. PMCID: PMC7322379.

46. Monteil V, Kwon H, Prado P, Hagelkrüys A, Wimmer RA, Stahl M, Leopoldi A, Garreta E, Hurtado Del Pozo C, Prosper F, Romero JP, Wirnsberger G, Zhang H, Slutsky AS, Conder R, Montserrat N, Mirazimi A, Penninger JM. 2020; Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 181:905–913.e7. DOI: 10.1016/j.cell.2020.04.004. PMID: 32333836. PMCID: PMC7181998.

47. Monteil V, Dyczynski M, Lauschke VM, Kwon H, Wirnsberger G, Youhanna S, Zhang H, Slutsky AS, Hurtado Del Pozo C, Horn M, Montserrat N, Penninger JM, Mirazimi A. 2020; Human soluble ACE2 improves the effect of remdesivir in SARS-CoV-2 infection. EMBO Mol Med. [Epub ahead of print]. DOI: 10.15252/emmm.202013426. PMID: 33179852. PMCID: PMC7799356.

48. Allison SJ. 2020; SARS-CoV-2 infection of kidney organoids prevented with soluble human ACE2. Nat Rev Nephrol. 16:316. DOI: 10.1038/s41581-020-0291-8. PMCID: PMC7187150. PMID: 32332922.

49. Ray K. 2020; Next-generation intestinal organoids. Nat Rev Gastroenterol Hepatol. 17:649. DOI: 10.1038/s41575-020-00371-8. PMID: 32994569.

50. Takebe T, Enomura M, Yoshizawa E, Kimura M, Koike H, Ueno Y, Matsuzaki T, Yamazaki T, Toyohara T, Osafune K, Nakauchi H, Yoshikawa HY, Taniguchi H. 2015; Vascularized and complex organ buds from diverse tissues via mesenchymal cell-driven condensation. Cell Stem Cell. 16:556–565. DOI: 10.1016/j.stem.2015.03.004. PMID: 25891906.

51. Saeidnia S, Manayi A, Abdollahi M. 2015; From in vitro experiments to in vivo and clinical studies; pros and cons. Curr Drug Discov Technol. 12:218–224. DOI: 10.2174/1570163813666160114093140. PMID: 26778084.

52. Bradley BT, Maioli H, Johnston R, Chaudhry I, Fink SL, Xu H, Najafian B, Deutsch G, Lacy JM, Williams T, Yarid N, Marshall DA. 2020; Histopathology and ultrastructural findings of fatal COVID-19 infections in Washington State: a case series. Lancet. 396:320–332. DOI: 10.1016/S0140-6736(20)31305-2. PMID: 32682491. PMCID: PMC7365650.

53. Park SE, Georgescu A, Huh D. 2019; Organoids-on-a-chip. Science. 364:960–965. DOI: 10.1126/science.aaw7894. PMID: 31171693. PMCID: PMC7764943.

54. Legallais C, Kim D, Mihaila SM, Mihajlovic M, Figliuzzi M, Bonandrini B, Salerno S, Yousef Yengej FA, Rookmaaker MB, Sanchez Romero N, Sainz-Arnal P, Pereira U, Pasqua M, Gerritsen KGF, Verhaar MC, Remuzzi A, Baptista PM, De Bartolo L, Masereeuw R, Stamatialis D. 2018; Bioengineering organs for blood detoxification. Adv Healthc Mater. 7:e1800430. DOI: 10.1002/adhm.201800430. PMID: 30230709.

55. Remuzzi A, Bonandrini B, Tironi M, Longaretti L, Figliuzzi M, Conti S, Zandrini T, Osellame R, Cerullo G, Raimondi MT. 2020; Effect of the 3D artificial nichoid on the morphology and mechanobiological response of mesenchymal stem cells cultured in vitro. Cells. 9:1873. DOI: 10.3390/cells9081873. PMID: 32796521. PMCID: PMC7464958.

Fig. 1
Several organoids have been established to study SARS-CoV-2. The upper refers to clinical symptoms of different organs infected by COVID-19, the medium is mainly emphasised on the mechanism of Inflammatory cytokine cascade after virus infection, and then, the bottom is now there are seveal organoids models (1. Brain organoids; 2. Heart organoids; 3. Lung organoids; 4. Gastrointestinal organoids; 5. Kidney organoids) used to screen corres-ponding anti-virual drugs, further-more, organoids-on-the-Chips is a more complete model of studying the entire process of virus infection in next days.
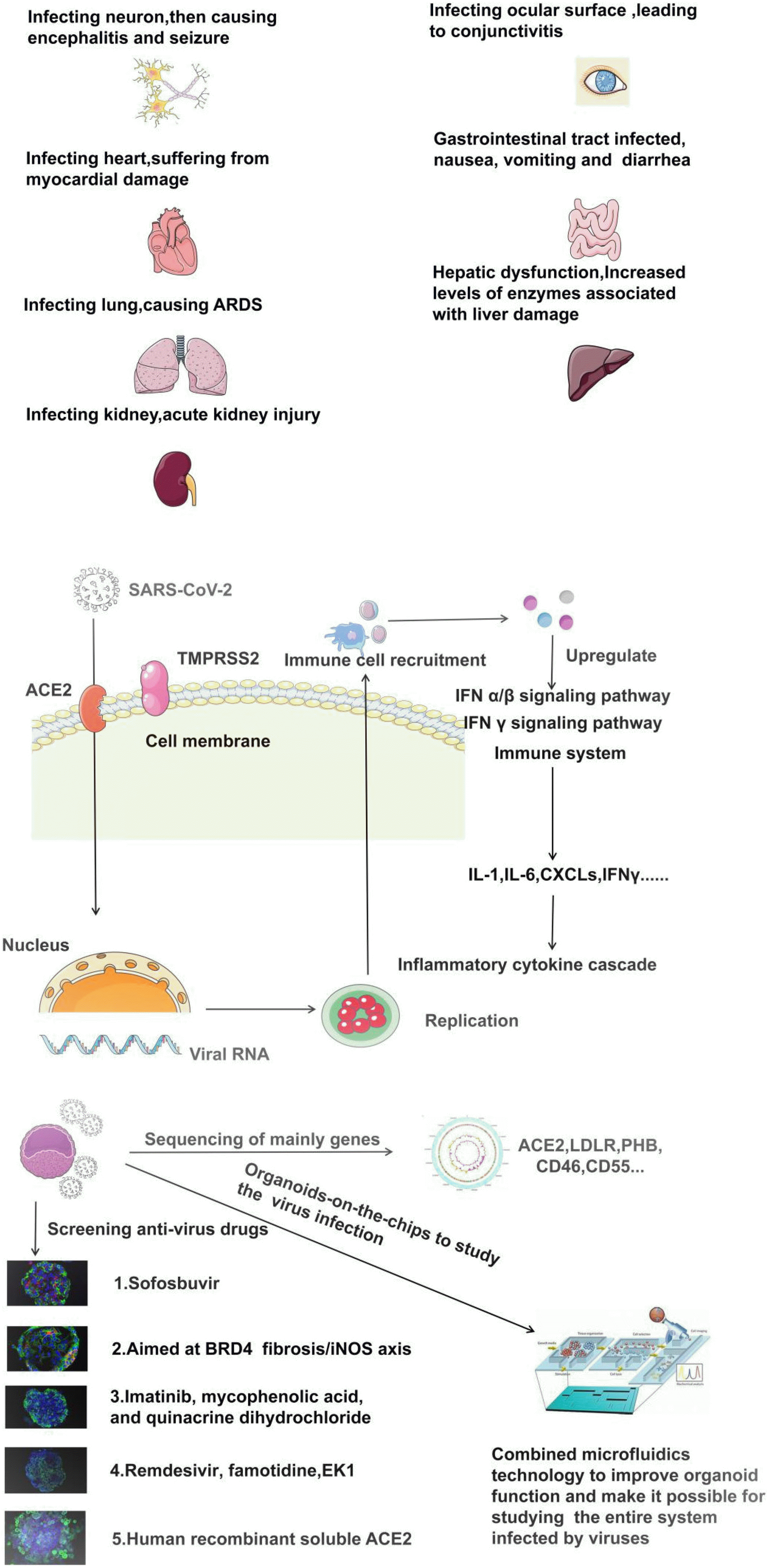
Table 1
Organoids’ cultural system for SARS-CoV-2 research




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download