Abstract
In Parkinson’s disease (PD) research, human neuroblastoma and immortalized neural cell lines have been widely used as in vitro models. The advancement in the field of reprogramming technology has provided tools for generating patient-specific induced pluripotent stem cells (hiPSCs) as well as human induced neuronal progenitor cells (hiNPCs). These cells have revolutionized the field of disease modeling, especially in neural diseases. Although the direct reprogramming to hiNPCs has several advantages over differentiation after hiPSC reprogramming, such as the time required and the simple procedure, relatively few studies have utilized hiNPCs. Here, we optimized the protocol for hiNPC reprogramming using pluripotency factors and Sendai virus. In addition, we generated hiNPCs of two healthy donors, a sporadic PD patient, and a familial patient with the LRRK2 G2019S mutation (L2GS). The four hiNPC cell lines are highly proliferative, expressed NPC markers, maintained the normal karyotype, and have the differentiation potential of dopaminergic neurons. Importantly, the patient hiNPCs show different apoptotic marker expression. Thus, these hiNPCs, in addition to hiPSCs, are a favorable option to study PD pathology.
Parkinson’s disease (PD) is the second most common neurodegenerative disorder after Alzheimer’s disease. Because the therapy and/or therapeutics for PD are still challenging, generating better models to recapitulate PD is imperative. Until now, the neuroblastoma cell line SH-SY5Y and immortalized Lund human mesencephalic (LUHMES) cells have been extensively used as in vitro models to study PD (1–3). These cell lines reproduce degeneration of dopaminergic neurons (DNs) and aggregation of α-synuclein in the presence of neurotoxin and exogenously introduced α-synuclein fibrils (1–3). However, since these cell lines originated from tumor or immortalized cells, they have limitations in representing the normal pathophysiology of PD. After the realization of “disease-in-a-dish” using human induced pluripotent stem cells (hiPSC) (4), for PD research, the use of hiPSC-derived neuronal progenitors (NPC) have advanced the understanding of pathology and efficacy testing of therapeutic agents (5). Currently hiPSC-derived NPC models are being continuously improved to optimize the efficiency of terminally differentiated dopaminergic neurons and to increase the expansion capacity of NPCs.
Besides the hiPSC-based disease model, the technology of direct reprogramming or transdifferentiation also has been adopted in disease modeling. Conversion of fibroblast to specific neurons shows impressive efficiency and phenotype (6–8). In 2010, induced neuron (iN) have been successfully converted from mouse fibroblasts by ectopic expression of three transcription factors—i.e., Brn2, Ascl1, and Myt1 (6). The iNs show electrophysiological currents in vitro, confirming functionally active neurons. In the past few years, there have been efforts to directly reprogram somatic cells into specific neuronal subtype including DN. To this end, iN-factor(s) and dopaminergic lineage transcription factors were introduced simultaneously (7–16). Through these methods, functional induced DN (iDN) that expressed TH and other midbrain DN markers have been generated from fibroblasts, blood cells (especially PBMC), or astrocytes (7–16). As shown in iN and iDN reprogramming, most of direct reprograming uses a set of transcription factors which are expressed in the target cells. However, there is another approach which use pluripotency factors for direct reprogramming (PDR). It is known that flexible intermediate cells are generated during PDR, and the intermediate cells are further converted into desired cells by target cell-specific environmental cues (17–29). Interestingly, direct reprogramming using just one pluripotent factor can generate expandable stem/progenitor cells (17, 21, 28). Because PDR generally use pluripotent factors, i.e., OCT4, SOX2, KLF4, and C-MYC, and the vector systems of iPSC reprogramming, researchers with experience in hiPSC reprogramming could follow PDR easily. Previously, we and others reported induced NPCs (iNPCs) by PDR (17, 21, 30, 31). The iNPCs showed stable expansion capacity, ease of differentiation, and long-term storage without any alteration in proliferation and differentiation potential (21, 31, 32). Recently, human iNPCs (hiNPCs) have been successfully differentiated into motor and dopaminergic neurons using specific patterning molecules (31). Therefore, hiNPCs, in addition to hiPSCs, are expected to provide another model for neural disease and drug discovery.
In this study, we optimized the protocol to generate hiNPC and described a sequential characterization procedure. Using our method, we successfully generated hiNPCs from a LRRK2 G2019S monogenic (L2GS) familial patient and a sporadic PD patient, in addition to hiNPCs from two healthy donors. Since these hiNPCs demonstrate the difference between normal and PD pathophysiology, we expect our cell lines will be excellent resources to model PD.
All human fibroblast cell lines used in this study were obtained from the Coriell Institute (USA). All information about the cell lines is summarized in Table 1. The cells were cultured in human fibroblast medium (MEM medium supplemented with 10% FBS, 1× sodium pyruvate, and 1× MEM-NEAA; Thermo Fisher Scientific, USA). For reprogramming purposes, the human fibroblasts were allowed exemption from IRB review by Public Institutional Review Board Designated by Ministry of Health and Welfare (P01-201802-31-001).
The reprogramming of fibroblasts to hiNPC was performed as previously described (21), with some modifications. Briefly, 30,000 human fibroblasts cells/well were plated onto Geltrex coated 24-well plates. The next day, human fibroblasts were transduced with Sendai virus (SeV) mixtures (CytoTune™-iPS 2.0 Sendai reprogramming kit, Thermo Fisher Scientific), according to the manufacturer’s instruction. After 24 h, cells were washed with Dulbecco’s Phosphate-Buffered Saline (DPBS, Welgene, Korea) and replaced with human neural reprogramming medium which consisted of 1:1 mixture of advanced DMEM/F-12 and Neurobasal medium, 0.05% AlbuMAX, 1× N2, 1× B27, 2 mM Glutamax, 0.11 mM 2-mercaptoethanol (Thermo Fisher Scientific), 3.0 μM CHIR99021 and 0.5 μM A83-01 (Tocris, UK), and 10 ng/ml hLIF (Peprotech, USA). The medium was replaced every other day. After seven days post transduction (dpt), growing cells were dissociated by Accutase (Millipore, USA) treatment and re-plated onto Geltrex coated 6-well plates. On 18~21 dpt, candidate colonies were manually picked and cultured in the coated plate with the same media for hiNPC reprogramming.
For PI staining, the hiNPCs were harvested using Accutase, and fixed with ice-cold 70% ethanol (Millipore) at 4°C overnight. The starting fibroblast cells were used as a control. Next, the cells were washed twice with DPBS, followed by incubation with solutions of 25 μg/ml RNase (Sigma-Aldrich, USA) and 10 μg/ml PI (Sigma-Aldrich) at 37°C for 30 min. The stained cells were analyzed using flow cytometry (BD Accuri® C6, BD Biosciences, USA). All data were exported to FCS files and analysed using the FlowJo software (ver. 10.5.3). To determine ploidy, hiNPCs labelled with PI were gated on single populations via FSC-A and FSC-H, to exclude doublets. Then, a histogram was generated to compare the intensities of the single cell population of hiNPCs.
The G2019S mutation in LRRK2 was confirmed by Sanger sequencing. Briefly, genomic DNA of hiNPCs and their parental fibroblasts were used as PCR templates. The amplified PCR products were sequenced and analyzed by Genotech (Daejeon, Korea). The primer sequences used in this experiment are listed in Supplementary Table S1.
hiNPCs were plated onto Geltrex-coated coverslips and supplemented with a neuronal differentiation medium, which was comprised of DMEM/F-12 (Thermo Fisher Scientific) medium supplemented with B27 without Vitamin A, 50 μg/ml 2-phospho-L-ascorbic acid (Sigma-Aldrich), 20 ng/ml BDNF and GDNF (Peprotech), and 0.5 mM dbcAMP (Enzo life science, USA). Half of the volume of total media was replaced every other day.
Immunocytochemistry was performed as described previously (21). Briefly, the cultured cells were fixed in 4% paraformaldehyde (Electron Microscopy Sciences, USA) for 10 min, followed by washing with DPBS. Next, the cells were blocked and permeabilized with 3% bovine serum albumin (BSA, Thermo Fisher Scientific) and 0.3% Triton X-100 (Sigma-Aldrich) in DPBS for 1 h at room temperature. All samples were then incubated with primary antibody solution overnight at 4°C. The next day, after washes with 0.1% BSA in DPBS, samples were incubated with Alexa Fluor 488- or Alexa Fluor 594-conjugated secondary antibodies (Thermo Fisher Scientific) for 1 h at room temperature. Images were captured using a Fluoview FV1000 confocal microscope (Olympus, Japan). The antibodies used in this experiment are listed in Supplementary Table S2.
Karyotyping of hiNPCs was conducted by Gendix (Seoul, Korea). A STR array was performed as previously described (33). Briefly, genomic DNA was extracted from hiNPCs and their parental fibroblast cells using a DNeasy Blood and Tissue kit (Qiagen, Germany), according to the manufacturer’s instructions. The STR array was analyzed by Humanpass (Seoul, Korea).
Detection of mycoplasma in cells was performed as previously described (34). Briefly, the cell pellets were collected by centrifugation, lysed at 55°C for 3 h, followed by 1 h incubation at 95°C with proteinase K (Sigma-Aldrich). PCR was performed using the extracted DNA as PCR templates. The primer sequences used in this experiment are listed in Supplementary Table S1.
RT-PCR analysis was performed as previously described (17). Total RNA was extracted using the RNeasy mini kit with a QIAshredder and DNase I (Qiagen) to avoid genomic DNA contamination. The RNA was reverse-transcribed using an iScript cDNA synthesis kit (Bio-Rad, USA), according to the manufacturer’s instructions. Next, the PCR reaction was performed using a 1:50 dilution of the cDNA template with an iQ SYBR Green supermix (Bio-Rad). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control gene. The primer sequences used in this experiment are listed in Supplementary Table S1.
Cells were treated with ice-cold sample lysis buffer consisting of 1% Triton X-100 (Sigma-Aldrich), 5 mM ethylenediaminetetraacetic acid (EDTA, Thermo Fischer Scientific), 1 mM phenylmethanesulfonylfluoride (PMSF, Thermo Fischer Scientific), and Xpert Protease Inhibitor Cocktail Solution (GenDEPOT, USA) in DPBS. Protein extracts were quantified with Protein Assay Dye Reagent Concentrate (Bio-Rad). An equal amount of total protein was separated by SDS-PAGE. All samples were then transferred to PVDF membrane (Bio-Rad) using a Wet/Tank Blotting System (Bio-Rad). The membranes were incubated first with blocking solution (Difco™ Skim milk, BD, USA), then primary antibodies were added, followed by the addition of horseradish peroxidase (HRP)-conjugated secondary antibodies (Cell Signaling Technology, USA). For detection of the oxidized signals from HRP, we added substrates (ECLTM Select Western Blotting Detection Reagent, GE Healthcare, USA). The HRP images of protein bands were acquired by a LAS-3000 Imager (Fujifilm, Japan). Primary antibodies used in this experiment are listed in Supplementary Table S2.
hiNPCs were seeded in 96-well plates, and 5 μM MG132 (Sigma-Aldrich) or equal volume of DMSO (Sigma-Aldrich) were added to the medium for 40 h. The viability of cells was measured using the EZ-Cytox viability assay kit (DoGenBio, Korea), according to the manufacturer’s instructions. Briefly, 10 μl EZ-Cytox were added to the medium. After 2.5 h, we measure the absorbance of the samples. We used the 450 nm of wavelength for measuring the absorbance and the 650 nm of wavelength for measuring the background.
We generated hiNPCs from fibroblasts of familial L2GS PD (FPD-hiNPC), sporadic PD (SPD-hiNPC) and two healthy donors (WT-hiNPC) by the PDR approach (Fig. 1a). After reprogramming, we manually picked some colonies and expanded them (Fig. 1b). To obtain intact hiNPC, we performed a step-wise analysis, as shown in Supplementary Fig. S1. Because we sometimes observed tetra ploidy in reprogrammed cells and previously aneuploid chromosomes often arise in reprogrammed cells (35), we first analyzed the ploidy of hiNPCs by simple PI staining-based flow cytometry to select diploid cells. We found that all established lines are diploid as the unreprogrammed starting fibroblasts (Fig. 2a). Second, we analyzed key markers of NPCs by immunocytochemistry, as shown in Fig. 2b. We selected the hiNPCs that expressed neural cadherin (N-CAD), PAX6, PLZF, and ZO1, as previously described (17, 21). Because almost all PAX6-expressing cells simultaneously expressed Ki-67, a cell cycle marker, we were able to confirm active proliferation of the cells as they were observed in culture. Because we wanted to confirm that the hiNPCs did not have the G2019S mutation in the LRRK2 gene (except the FPD-hiNPC), we sequenced the genomic locus of LRRK2 G2019S in the starting fibroblasts and corresponding hiNPC lines. The substitution from G to A was observed in one allele of FPD-hiNPCs and its parental fibroblasts (Fig. 2c). The SPD-hiNPCs, and WT1- and WT2-hiNPCs contained the normal nucleotide sequence in the LRRK2 G2019 locus (Fig. 2c). Thus, the isolated hiNPC candidates have diploid chromosomes, express key NPC markers, and maintain the genetic background.
Next, we checked the differentiation potential of hiNPC candidates. We differentiated the cells spontaneously using neuronal differentiation medium. We observed that neurite outgrowths started within 3 days, and long and arborized neurites were observed after 21 days of differentiation (Fig. 1b). Because we sought to use the hiNPCs as a PD model, differentiation to dopaminergic neurons (DN) are a critical characteristic. We observed that tyrosine hydroxylase (TH) was co-stained with a pan-neuronal marker (TUJ1) in the differentiated cells from all hiNPCs. We also observed an astrocyte marker, GFAP, and a mature neuronal marker, MAP2, after 21 days of differentiation (Fig. 2d). Consistent with the immunocytochemistry results, the mRNA expression of mature neuronal markers such as MAP2, NEUN, and SYNAPSIN1 were also increased in differentiated cells compared to that of undifferentiated hiNPCs (Fig. 2e). To gain insight into neuronal function, we further examined the expression of neurotransmitter receptors. Glutamate receptors are known to be activated in normal physiology by mediating excitatory synaptic transmission in the nervous system (36). We found that the mRNA levels of GRIN1 (NMDA receptor) and GRIA2 (AMPA receptor) were greatly increased in differentiated cells, expecting active neurotransmitter function (Fig. 2e). Further, the expression of S100B, one of the glial markers, was also increased after differentiation (Fig. 2e). Thus, we confirmed the multipotency of hiNPC candidates and the functionality of differentiated neurons indirectly.
After characterization and confirmation of biological functions of the reprogrammed cells, we tested several basic requirements before cryopreservation. Because chromosomal anomaly often occurs in stem cells (37), karyotyping and G-banding analysis are useful to promptly check the genomic integrity. We found that all hiNPCs maintained a normal karyotype after culturing over eight passages (Fig. 3a). Because we usually reprogram multiple independent cells in a single batch experiment, contamination of other cells could be a potential risk. Thus, short tandem repeat (STR) analysis is important and always required after reprogramming and before banking to confirm clonality of the reprogrammed cells. We confirmed that all hiNPCs showed the same STR profile as that of the parental fibroblasts respectively (Fig. 3b). Finally, we confirmed the absence of mycoplasma by PCR amplification of the specific rRNA region of mycoplasma (Fig. 3c) (34). As above, we checked the characteristics of hiNPCs using NPC markers, differentiation potential and neuronal functionality after spontaneous differentiation using neuronal markers. To determine if the hiNPCs were adequate for long-term storage, the integrity of the reprogrammed cells was also checked. In our experience, we could efficiently analyze the hiNPC candidates with minimal effort by the proposed characterization flowchart (Supplementary Fig. S1).
To establish the cellular model of PD, various stress reagents have been widely used to mimic the vulnerability of DNs (38–40). One of the proteasome inhibitors, MG132, impairs the intracellular protein clearance system such as the ubiquitin proteasome system, resulting in cell death (41, 42). Because PD patient-derived cells showed more severe cell death by proteasomal stress than healthy controls, proteasome inhibitors has been widely used for PD modeling (39, 40, 42). To confirm whether our hiNPCs represent a PD phenotype such as apoptosis, we treated them with MG132 or DMSO (Fig. 4a). As expected, we found more apoptotic cells in FPD-hiNPCs and surprisingly in SPD-hiNPCs than in WT-hiNPCs by MG132 treatment whereas we did not observe the difference of cell death in all DMSO controls (Fig. 4b). To quantify cell death, we performed WST based cell viability assay and immunoblot for cleaved CASPASE3 (cCASP3). Consistent with the morphology of cells, when the DMSO control was set at 100% in each cell line respectively, FPD- and SPD-hiNPCs exhibited significantly decreased cell viability (27.5±0.4% and 30.4±2.1% respectively) than WT1-, WT2-hiNPCs (39.1±1.0% and 38.8±1.1% respectively) by MG132 treatment (Fig. 4c). We also confirmed increased expression of cCASP3 in FPD- and SPD-hiNPCs relative to WT1- and WT2-hiNPCs by MG132 treatment (Fig. 4d, 4e). These results demonstrate that PD patients-derived hiNPCs are more sensitive to the proteasome stress than healthy controls, consequently resulting in more cell death in hiNPCs derived from PD patients than hiNPCs derived from healthy donors. Thus, our hiNPCs are useful to model both familial and sporadic PD and could be used to develop various other PD modeling paradigms.
Here, we optimized a direct reprogramming protocol for hiNPCs and proposed a step-wise characterization process. We also showed that hiNPCs are adequate and acceptable resources for an in vitro cellular model for PD. We expect that directly reprogrammed hiNPCs could be used for other neural diseases, and would be better or equivalent to iPSC-derived models.
Acknowledgments
This work was supported by the KRIBB Research Initiative and Stem Cell Research Program through the National Research Foundation of Korea funded by the Ministry of Science and ICT (2013M3A9B4076483, 2015M3A9C7030128, 2018M3A9H3023077, and 2016K1A3A1A61006001) and a grant from Ministry of Food and Drug Safety in 2018 (18172MFDS182).
Notes
Supplementary Materials
Supplementary data including two tables and one figure can be found with this article online at http://pdf.medrang.co.kr/paper/pdf/IJSC/IJSC-12-s19075.pdf.
References
1. Falkenburger BH, Saridaki T, Dinter E. Cellular models for Parkinson’s disease. J Neurochem. 2016; 139(Suppl 1):121–130. DOI: 10.1111/jnc.13618. PMID: 27091001.

2. Lotharius J, Falsig J, van Beek J, Payne S, Dringen R, Brundin P, Leist M. Progressive degeneration of human mesencephalic neuron-derived cells triggered by dopamine-dependent oxidative stress is dependent on the mixed-lineage kinase pathway. J Neurosci. 2005; 25:6329–6342. DOI: 10.1523/JNEUROSCI.1746-05.2005. PMID: 16000623. PMCID: PMC6725277.

3. Jang W, Kim HJ, Li H, Jo KD, Lee MK, Yang HO. The neuroprotective effect of erythropoietin on rotenone-induced neurotoxicity in SH-SY5Y cells through the induction of autophagy. Mol Neurobiol. 2016; 53:3812–3821. DOI: 10.1007/s12035-015-9316-x. PMID: 26156288.

4. Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007; 131:861–872. DOI: 10.1016/j.cell.2007.11.019. PMID: 18035408.

5. Torrent R, De Angelis Rigotti F, Dell’Era P, Memo M, Raya A, Consiglio A. Using iPS cells toward the understanding of Parkinson’s disease. J Clin Med. 2015; 4:548–566. DOI: 10.3390/jcm4040548. PMID: 26239346. PMCID: PMC4470155.

6. Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010; 463:1035–1041. DOI: 10.1038/nature08797. PMID: 20107439. PMCID: PMC2829121.

7. Yoo J, Lee E, Kim HY, Youn DH, Jung J, Kim H, Chang Y, Lee W, Shin J, Baek S, Jang W, Jun W, Kim S, Hong J, Park HJ, Lengner CJ, Moh SH, Kwon Y, Kim J. Electromagnetized gold nanoparticles mediate direct lineage reprogramming into induced dopamine neurons in vivo for Parkinson’s disease therapy. Nat Nanotechnol. 2017; 12:1006–1014. DOI: 10.1038/nnano.2017.133. PMID: 28737745.

8. Pfisterer U, Kirkeby A, Torper O, Wood J, Nelander J, Dufour A, Björklund A, Lindvall O, Jakobsson J, Parmar M. Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci U S A. 2011; 108:10343–10348. DOI: 10.1073/pnas.1105135108. PMID: 21646515. PMCID: PMC3121829.

9. Caiazzo M, Dell’Anno MT, Dvoretskova E, Lazarevic D, Taverna S, Leo D, Sotnikova TD, Menegon A, Roncaglia P, Colciago G, Russo G, Carninci P, Pezzoli G, Gainetdinov RR, Gustincich S, Dityatev A, Broccoli V. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011; 476:224–227. DOI: 10.1038/nature10284. PMID: 21725324.

10. Addis RC, Hsu FC, Wright RL, Dichter MA, Coulter DA, Gearhart JD. Efficient conversion of astrocytes to functional midbrain dopaminergic neurons using a single polycistronic vector. PLoS One. 2011; 6:e28719. DOI: 10.1371/journal.pone.0028719. PMID: 22174877. PMCID: PMC3235158.

11. Liu X, Li F, Stubblefield EA, Blanchard B, Richards TL, Larson GA, He Y, Huang Q, Tan AC, Zhang D, Benke TA, Sladek JR, Zahniser NR, Li CY. Direct reprogramming of human fibroblasts into dopaminergic neuron-like cells. Cell Res. 2012; 22:321–332. DOI: 10.1038/cr.2011.181. PMID: 22105488. PMCID: PMC3271588.

12. Kim J, Su SC, Wang H, Cheng AW, Cassady JP, Lodato MA, Lengner CJ, Chung CY, Dawlaty MM, Tsai LH, Jaenisch R. Functional integration of dopaminergic neurons directly converted from mouse fibroblasts. Cell Stem Cell. 2011; 9:413–419. DOI: 10.1016/j.stem.2011.09.011. PMID: 22019014. PMCID: PMC3210333.

13. Torper O, Pfisterer U, Wolf DA, Pereira M, Lau S, Jakobsson J, Björklund A, Grealish S, Parmar M. Generation of induced neurons via direct conversion in vivo. Proc Natl Acad Sci U S A. 2013; 110:7038–7043. DOI: 10.1073/pnas.1303829110. PMID: 23530235. PMCID: PMC3637783.

14. Jiang H, Xu Z, Zhong P, Ren Y, Liang G, Schilling HA, Hu Z, Zhang Y, Wang X, Chen S, Yan Z, Feng J. Cell cycle and p53 gate the direct conversion of human fibroblasts to dopaminergic neurons. Nat Commun. 2015; 6:10100. DOI: 10.1038/ncomms10100. PMID: 26639555. PMCID: PMC4672381.

15. Park H, Kim H, Yoo J, Lee J, Choi H, Baek S, Lee CJ, Kim J, Lengner CJ, Sung JS, Kim J. Homogeneous generation of iDA neurons with high similarity to bona fide DA neurons using a drug inducible system. Biomaterials. 2015; 72:152–162. DOI: 10.1016/j.biomaterials.2015.09.002. PMID: 26370928.

16. Rivetti di Val Cervo P, Romanov RA, Spigolon G, Masini D, Martín-Montañez E, Toledo EM, La Manno G, Feyder M, Pifl C, Ng YH, Sánchez SP, Linnarsson S, Wernig M, Harkany T, Fisone G, Arenas E. Induction of functional dopamine neurons from human astrocytes in vitro and mouse astrocytes in a Parkinson’s disease model. Nat Biotechnol. 2017; 35:444–452. DOI: 10.1038/nbt.3835. PMID: 28398344.

17. Kim J, Efe JA, Zhu S, Talantova M, Yuan X, Wang S, Lipton SA, Zhang K, Ding S. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc Natl Acad Sci U S A. 2011; 108:7838–7843. DOI: 10.1073/pnas.1103113108. PMID: 21521790. PMCID: PMC3093517.

18. Efe JA, Hilcove S, Kim J, Zhou H, Ouyang K, Wang G, Chen J, Ding S. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat Cell Biol. 2011; 13:215–222. DOI: 10.1038/ncb2164. PMID: 21278734.

19. Thier M, Wörsdörfer P, Lakes YB, Gorris R, Herms S, Opitz T, Seiferling D, Quandel T, Hoffmann P, Nöthen MM, Brüstle O, Edenhofer F. Direct conversion of fibroblasts into stably expandable neural stem cells. Cell Stem Cell. 2012; 10:473–479. DOI: 10.1016/j.stem.2012.03.003. PMID: 22445518.

20. Kim J, Ambasudhan R, Ding S. Direct lineage reprogramming to neural cells. Curr Opin Neurobiol. 2012; 22:778–784. DOI: 10.1016/j.conb.2012.05.001. PMID: 22652035. PMCID: PMC4945246.

21. Zhu S, Ambasudhan R, Sun W, Kim HJ, Talantova M, Wang X, Zhang M, Zhang Y, Laurent T, Parker J, Kim HS, Zaremba JD, Saleem S, Sanz-Blasco S, Masliah E, McKercher SR, Cho YS, Lipton SA, Kim J, Ding S. Small molecules enable OCT4-mediated direct reprogramming into expandable human neural stem cells. Cell Res. 2014; 24:126–129. DOI: 10.1038/cr.2013.156. PMID: 24296783. PMCID: PMC3879704.

22. Zhu S, Russ HA, Wang X, Zhang M, Ma T, Xu T, Tang S, Hebrok M, Ding S. Human pancreatic beta-like cells converted from fibroblasts. Nat Commun. 2016; 7:10080. DOI: 10.1038/ncomms10080. PMID: 26733021. PMCID: PMC4729817.

23. Wang H, Cao N, Spencer CI, Nie B, Ma T, Xu T, Zhang Y, Wang X, Srivastava D, Ding S. Small molecules enable cardiac reprogramming of mouse fibroblasts with a single factor, Oct4. Cell Rep. 2014; 6:951–960. DOI: 10.1016/j.celrep.2014.01.038. PMID: 24561253. PMCID: PMC4004339.

24. Lee JH, Mitchell RR, McNicol JD, Shapovalova Z, Laronde S, Tanasijevic B, Milsom C, Casado F, Fiebig-Comyn A, Collins TJ, Singh KK, Bhatia M. Single transcription factor conversion of human blood fate to NPCs with CNS and PNS developmental capacity. Cell Rep. 2015; 11:1367–1376. DOI: 10.1016/j.celrep.2015.04.056. PMID: 26004181.

25. Kurian L, Sancho-Martinez I, Nivet E, Aguirre A, Moon K, Pendaries C, Volle-Challier C, Bono F, Herbert JM, Pulecio J, Xia Y, Li M, Montserrat N, Ruiz S, Dubova I, Rodriguez C, Denli AM, Boscolo FS, Thiagarajan RD, Gage FH, Loring JF, Laurent LC, Izpisua Belmonte JC. Conversion of human fibroblasts to angioblast-like progenitor cells. Nat Methods. 2013; 10:77–83. DOI: 10.1038/nmeth.2255. PMID: 23202434. PMCID: PMC3531579.

26. Zhu S, Rezvani M, Harbell J, Mattis AN, Wolfe AR, Benet LZ, Willenbring H, Ding S. Mouse liver repopulation with hepatocytes generated from human fibroblasts. Nature. 2014; 508:93–97. DOI: 10.1038/nature13020. PMID: 24572354. PMCID: PMC4161230.

27. Li K, Zhu S, Russ HA, Xu S, Xu T, Zhang Y, Ma T, Hebrok M, Ding S. Small molecules facilitate the reprogramming of mouse fibroblasts into pancreatic lineages. Cell Stem Cell. 2014; 14:228–236. DOI: 10.1016/j.stem.2014.01.006. PMID: 24506886. PMCID: PMC4747235.

28. Zhang Y, Cao N, Huang Y, Spencer CI, Fu JD, Yu C, Liu K, Nie B, Xu T, Li K, Xu S, Bruneau BG, Srivastava D, Ding S. Expandable cardiovascular progenitor cells reprogrammed from fibroblasts. Cell Stem Cell. 2016; 18:368–381. DOI: 10.1016/j.stem.2016.02.001. PMID: 26942852. PMCID: PMC5826660.

29. Lu J, Liu H, Huang CT, Chen H, Du Z, Liu Y, Sherafat MA, Zhang SC. Generation of integration-free and region-specific neural progenitors from primate fibroblasts. Cell Rep. 2013; 3:1580–1591. DOI: 10.1016/j.celrep.2013.04.004. PMID: 23643533. PMCID: PMC3786191.

30. Han DW, Tapia N, Hermann A, Hemmer K, Höing S, Araúzo-Bravo MJ, Zaehres H, Wu G, Frank S, Moritz S, Greber B, Yang JH, Lee HT, Schwamborn JC, Storch A, Schöler HR. Direct reprogramming of fibroblasts into neural stem cells by defined factors. Cell Stem Cell. 2012; 10:465–472. DOI: 10.1016/j.stem.2012.02.021. PMID: 22445517.

31. Sheng C, Jungverdorben J, Wiethoff H, Lin Q, Flitsch LJ, Eckert D, Hebisch M, Fischer J, Kesavan J, Weykopf B, Schneider L, Holtkamp D, Beck H, Till A, Wüllner U, Ziller MJ, Wagner W, Peitz M, Brüstle O. A stably self-renewing adult blood-derived induced neural stem cell exhibiting patternability and epigenetic rejuvenation. Nat Commun. 2018; 9:4047. DOI: 10.1038/s41467-018-06398-5. PMID: 30279449. PMCID: PMC6168501.

32. Cairns DM, Chwalek K, Moore YE, Kelley MR, Abbott RD, Moss S, Kaplan DL. Expandable and rapidly differentiating human induced neural stem cell lines for multiple tissue engineering applications. Stem Cell Reports. 2016; 7:557–570. DOI: 10.1016/j.stemcr.2016.07.017. PMID: 27569063. PMCID: PMC5032182.

33. Jung KB, Lee H, Son YS, Lee JH, Cho HS, Lee MO, Oh JH, Lee J, Kim S, Jung CR, Kim J, Son MY. In vitro and in vivo imaging and tracking of intestinal organoids from human induced pluripotent stem cells. FASEB J. 2018; 32:111–122. DOI: 10.1096/fj.201700504R. PMID: 28855280.

34. Tang J, Hu M, Lee S, Roblin R. A polymerase chain reaction based method for detecting Mycoplasma/Acholeplasma contaminants in cell culture. J Microbiol Methods. 2000; 39:121–126. DOI: 10.1016/S0167-7012(99)00107-4. PMID: 10576701.

35. Weissbein U, Ben-David U, Benvenisty N. Virtual karyotyping reveals greater chromosomal stability in neural cells derived by transdifferentiation than those from stem cells. Cell Stem Cell. 2014; 15:687–691. DOI: 10.1016/j.stem.2014.10.018. PMID: 25479746.

36. Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010; 62:405–496. DOI: 10.1124/pr.109.002451. PMID: 20716669. PMCID: PMC2964903.

37. Potapova TA, Zhu J, Li R. Aneuploidy and chromosomal instability: a vicious cycle driving cellular evolution and cancer genome chaos. Cancer Metastasis Rev. 2013; 32:377–389. DOI: 10.1007/s10555-013-9436-6. PMID: 23709119. PMCID: PMC3825812.

38. Nguyen HN, Byers B, Cord B, Shcheglovitov A, Byrne J, Gujar P, Kee K, Schüle B, Dolmetsch RE, Langston W, Palmer TD, Pera RR. LRRK2 mutant iPSC-derived DA neurons demonstrate increased susceptibility to oxidative stress. Cell Stem Cell. 2011; 8:267–280. DOI: 10.1016/j.stem.2011.01.013. PMID: 21362567. PMCID: PMC3578553.

39. Cooper O, Seo H, Andrabi S, Guardia-Laguarta C, Graziotto J, Sundberg M, McLean JR, Carrillo-Reid L, Xie Z, Osborn T, Hargus G, Deleidi M, Lawson T, Bogetofte H, Perez-Torres E, Clark L, Moskowitz C, Mazzulli J, Chen L, Volpicelli-Daley L, Romero N, Jiang H, Uitti RJ, Huang Z, Opala G, Scarffe LA, Dawson VL, Klein C, Feng J, Ross OA, Trojanowski JQ, Lee VM, Marder K, Surmeier DJ, Wszolek ZK, Przedborski S, Krainc D, Dawson TM, Isacson O. Pharmacological rescue of mitochondrial deficits in iPSC-derived neural cells from patients with familial Parkinson’s disease. Sci Transl Med. 2012; 4:141ra90. DOI: 10.1126/scitranslmed.3003985. PMID: 22764206. PMCID: PMC3462009.

40. Liu GH, Qu J, Suzuki K, Nivet E, Li M, Montserrat N, Yi F, Xu X, Ruiz S, Zhang W, Wagner U, Kim A, Ren B, Li Y, Goebl A, Kim J, Soligalla RD, Dubova I, Thompson J, Yates J 3rd, Esteban CR, Sancho-Martinez I, Izpisua Belmonte JC. Progressive degeneration of human neural stem cells caused by pathogenic LRRK2. Nature. 2012; 491:603–607. DOI: 10.1038/nature11557. PMID: 23075850. PMCID: PMC3504651.

41. Caldwell KA, Tucci ML, Armagost J, Hodges TW, Chen J, Memon SB, Blalock JE, DeLeon SM, Findlay RH, Ruan Q, Webber PJ, Standaert DG, Olson JB, Caldwell GA. Investigating bacterial sources of toxicity as an environmental contributor to dopaminergic neurodegeneration. PLoS One. 2009; 4:e7227. DOI: 10.1371/journal.pone.0007227. PMID: 19806188. PMCID: PMC2751819.

42. Bentea E, Verbruggen L, Massie A. The proteasome inhibition model of Parkinson’s disease. J Parkinsons Dis. 2017; 7:31–63. DOI: 10.3233/JPD-160921. PMID: 27802243. PMCID: PMC5302045.
Fig. 1
Direct reprogramming to generate hiNPCs. (a) Schematic diagram to show direct reprogramming of fibroblasts to hiNPCs. (b) Representative bright field images of fibroblasts, a reprogrammed hiNPC colony, clonally expanded hiNPCs, and spontaneously differentiated cells from hiNPCs. Scale bars represent 100 μm.
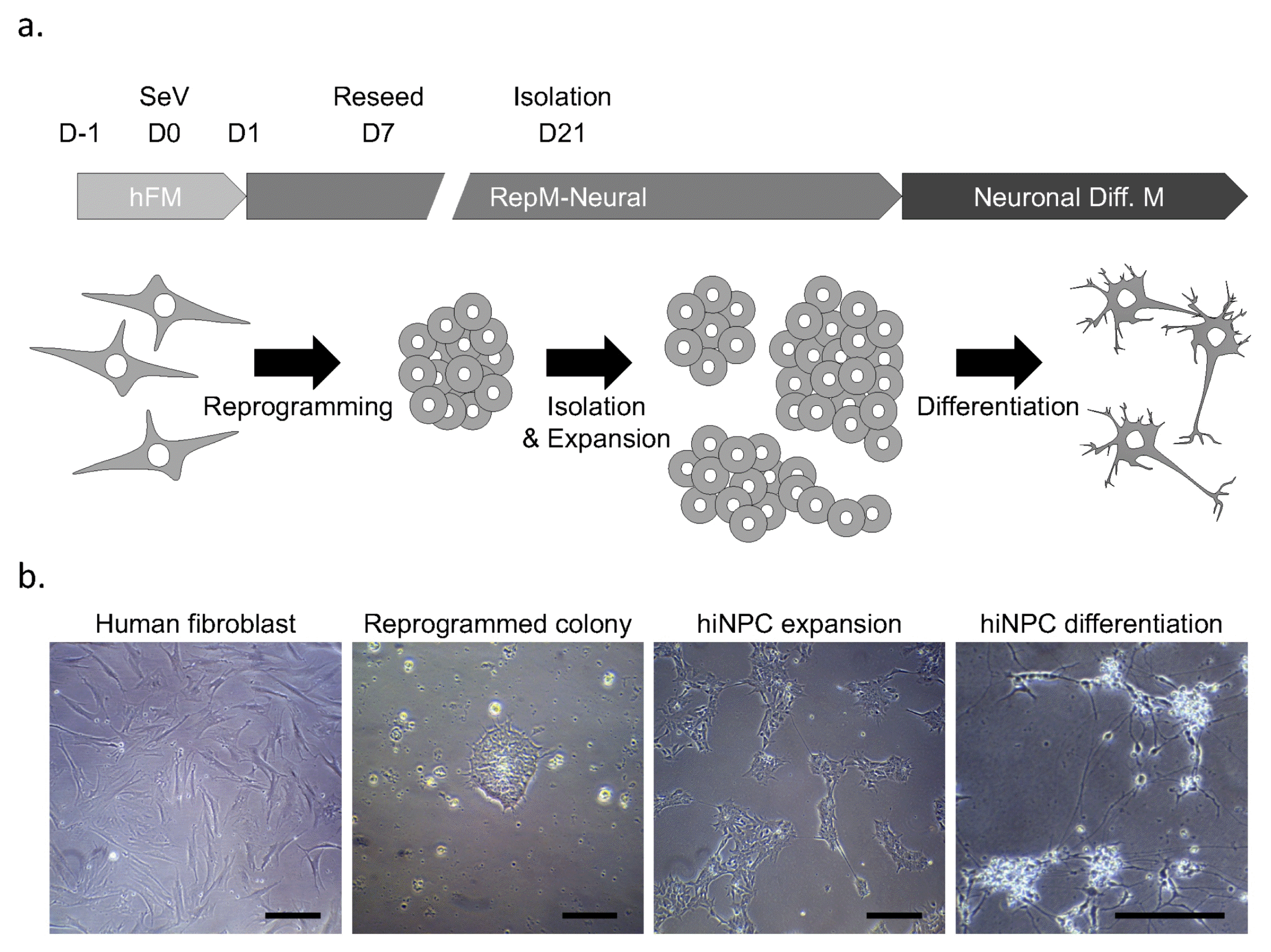
Fig. 2
Characterization of hiNPC lines. (a) Flow cytometry to detect ploidy of PI stained hiNPCs. Human fibroblasts from healthy donors were used as a 2n control. WT1, WT2, FPD, and SPD represent AG02261-hiNPC, GM01680-hiNPC, ND38262-hiNPC, and AG20446-hiNPC, respectively. (b) Immunocytochemistry for key NPC markers in hiNPCs. Ho. represents Hoechst33342 for staining nuclei. Scale bars represent 50 μm. (c) Mutation analysis of generated hiNPCs and the parental fibroblasts. The arrow indicates the G2019S mutation site in LRRK2. The red arrow indicates heterozygosity of G and A. (d) Immunocytochemistry of differentiated cells from hiNPCs with representative markers for pan-neurons, dopaminergic neurons, mature neurons, and glia. All hiNPCs were differentiated for 21 days. Scale bars represent 50 μm. (e) mRNA expression of MAP2, NEUN, SYNAPSIN1, GRIN1, GRIA2, and S100B in undifferentiated and differentiated hiNPCs.
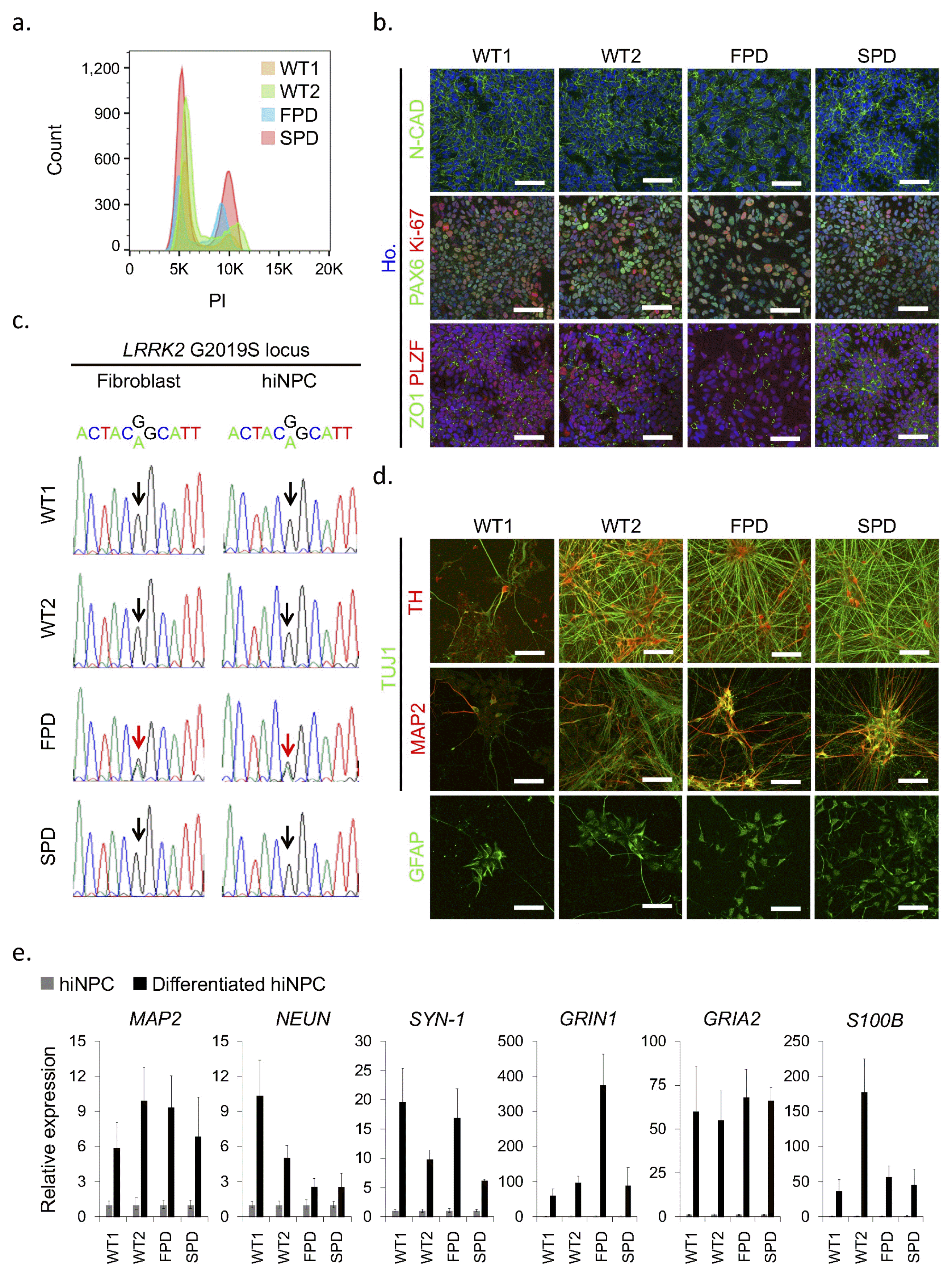
Fig. 3
Quality check of hiNPC lines before cryopreservation. (a) Karyotypes of established hiNPC lines at passage 9, 13, 8, and 13 of WT1-, WT2-, FPD-, and SPD-hiNPC, respectively. (b) STR analysis comparing starting fibroblasts and their corresponding hiNPCs. (c) Mycoplasma test by PCR. A 100 bp ladder was used.
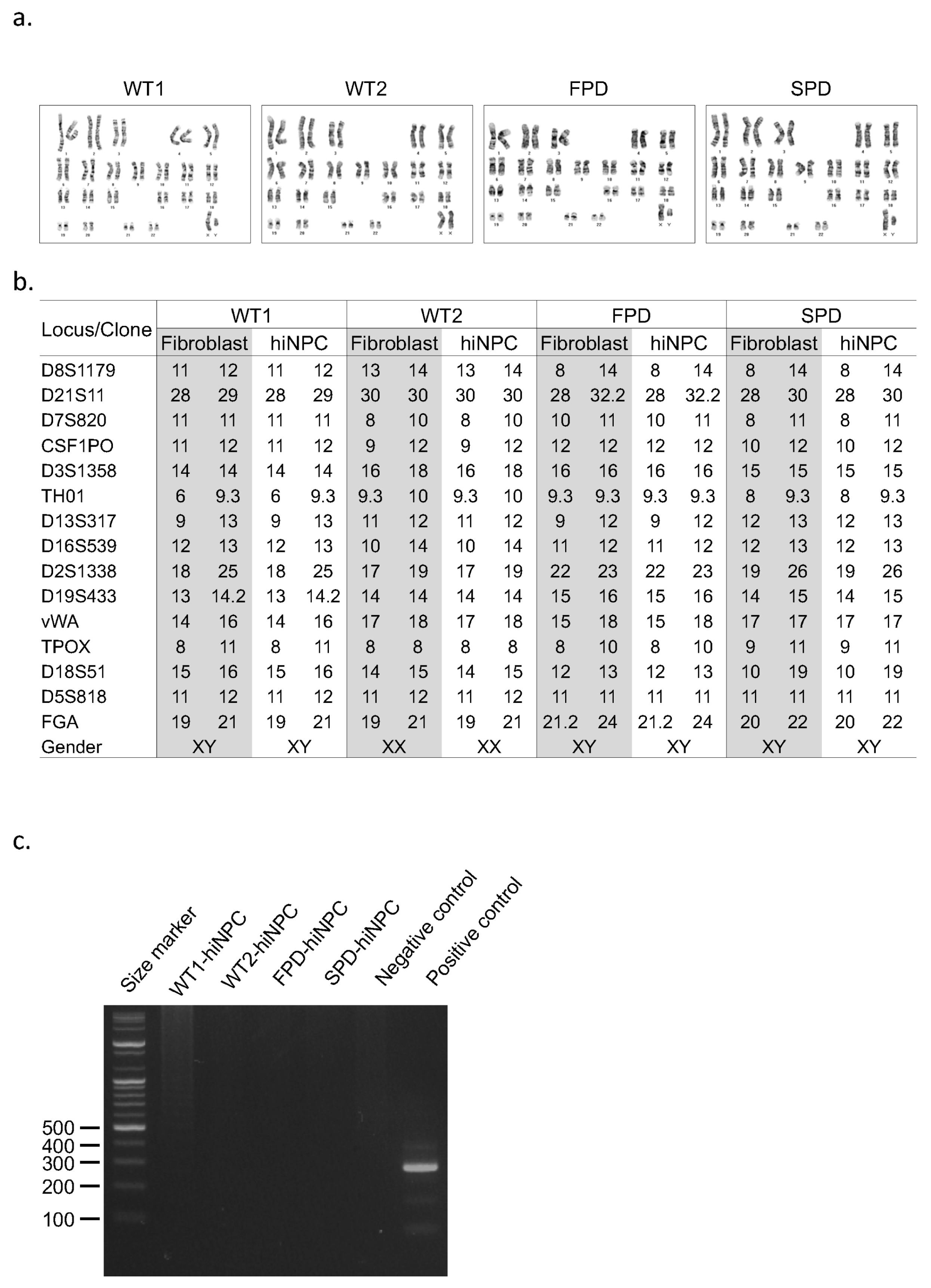
Fig. 4
hiNPCs as a PD model. (a) Schematic diagram for PD modeling. (b) Representative bright field images of hiNPCs after treatment with MG132. Scale bars represent 200 μm. (c) WST based cell viability assay with DMSO or MG132 treatment. All values indicate relative level of its corresponding DMSO control groups. (d) Immunoblot of cCASP3 in hiNPCs with or without MG132 treatment. GAPDH was used as an internal control. (e) Quantification of the band intensities. All values indicate relative level of cCASP3 to GAPDH. ** represents p<0.01; *** represents p<0.001 using Student’s t-test.
Table 1
Lists of human fibroblasts used in this study




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download