Introduction
Overview of the procedure
Fig. 1
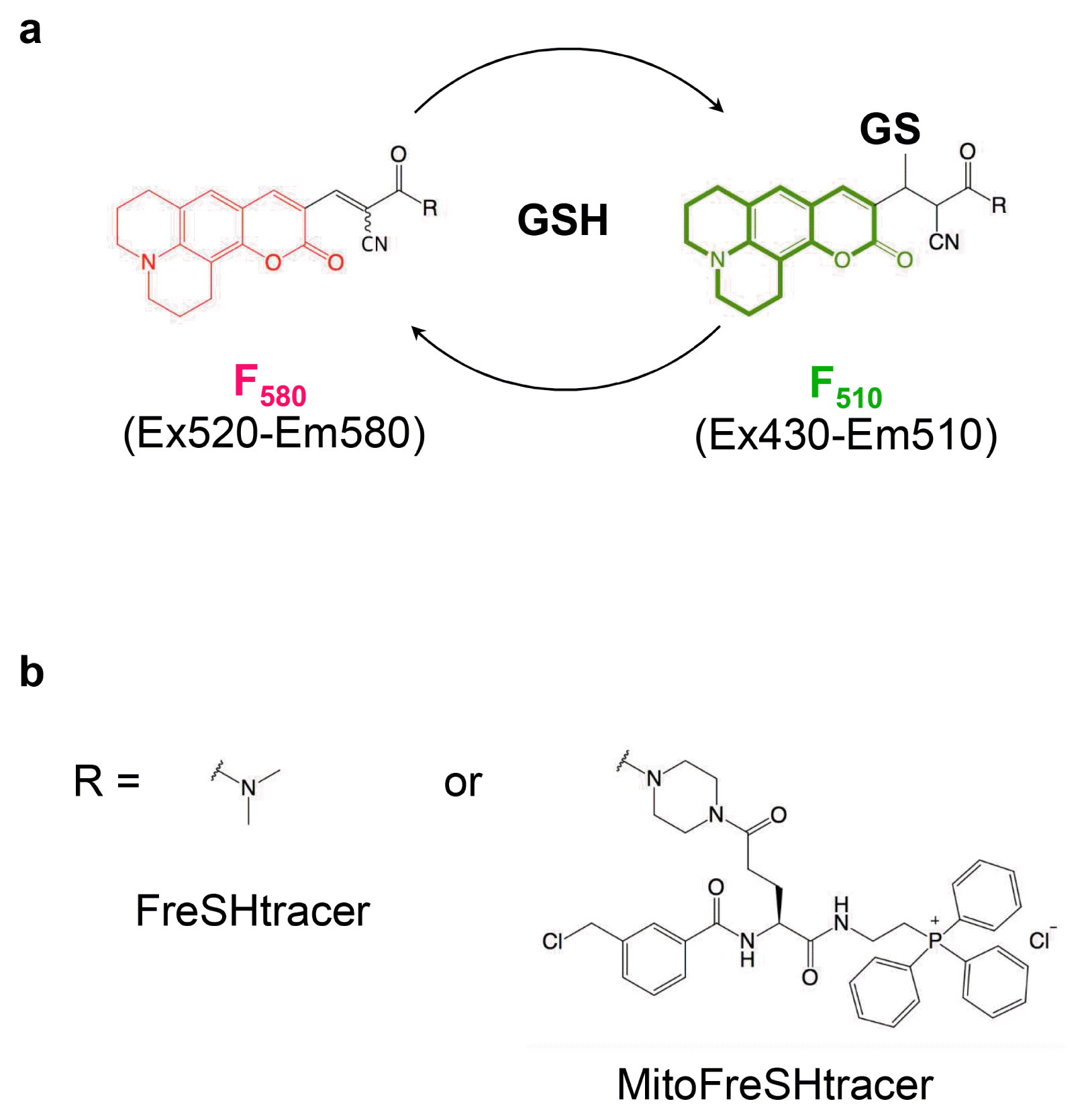
Fig. 2
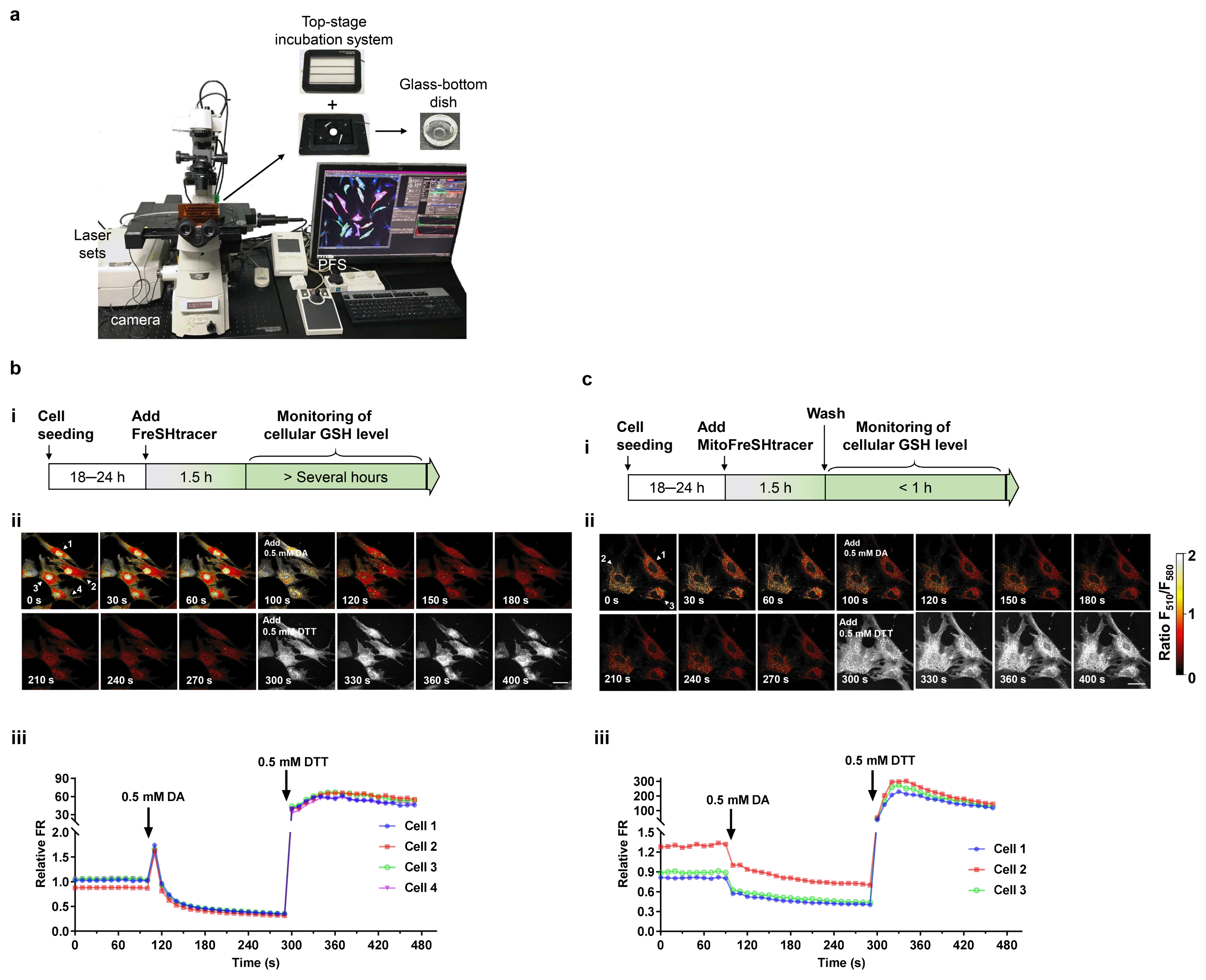
Fig. 3
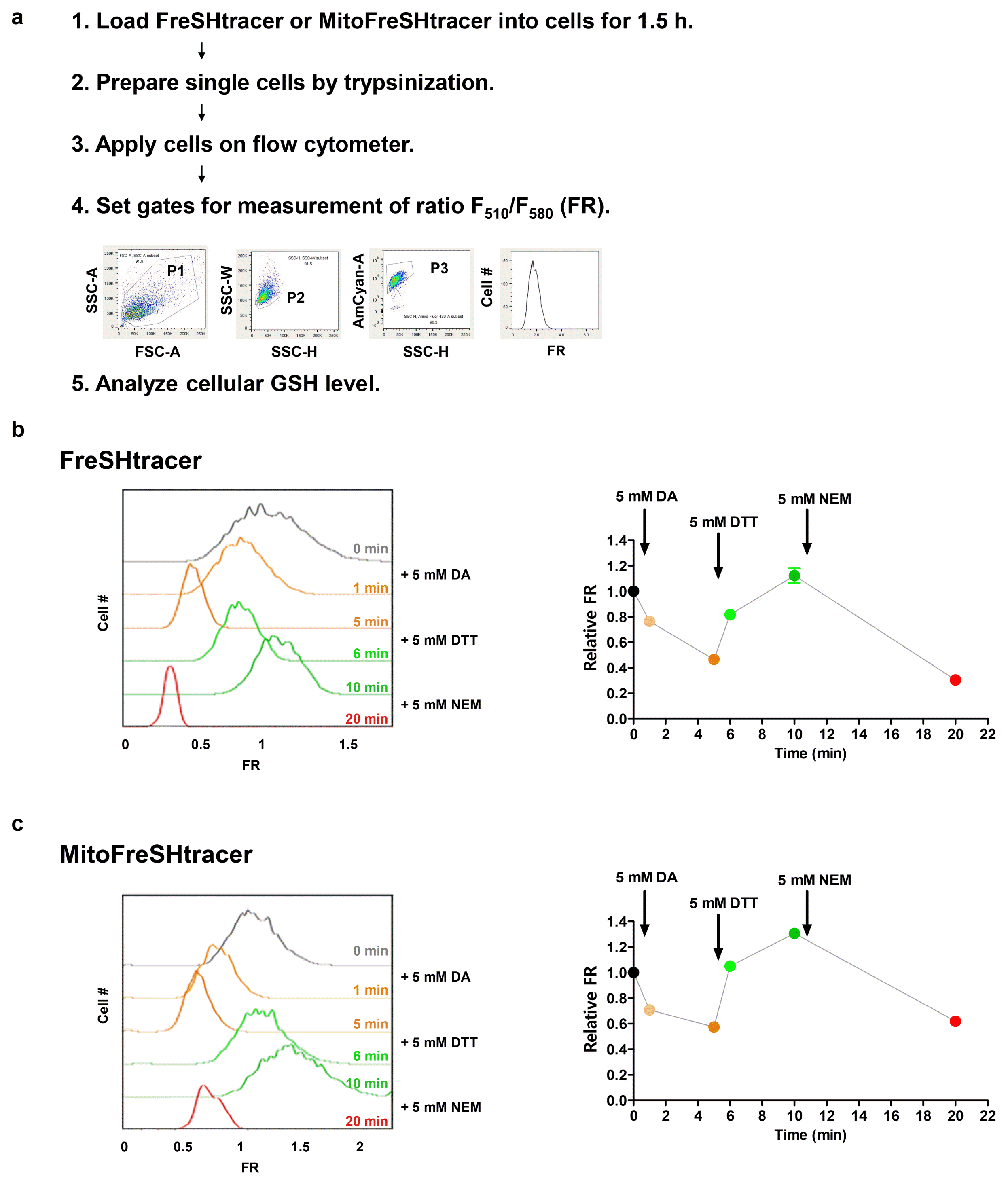
Fig. 4
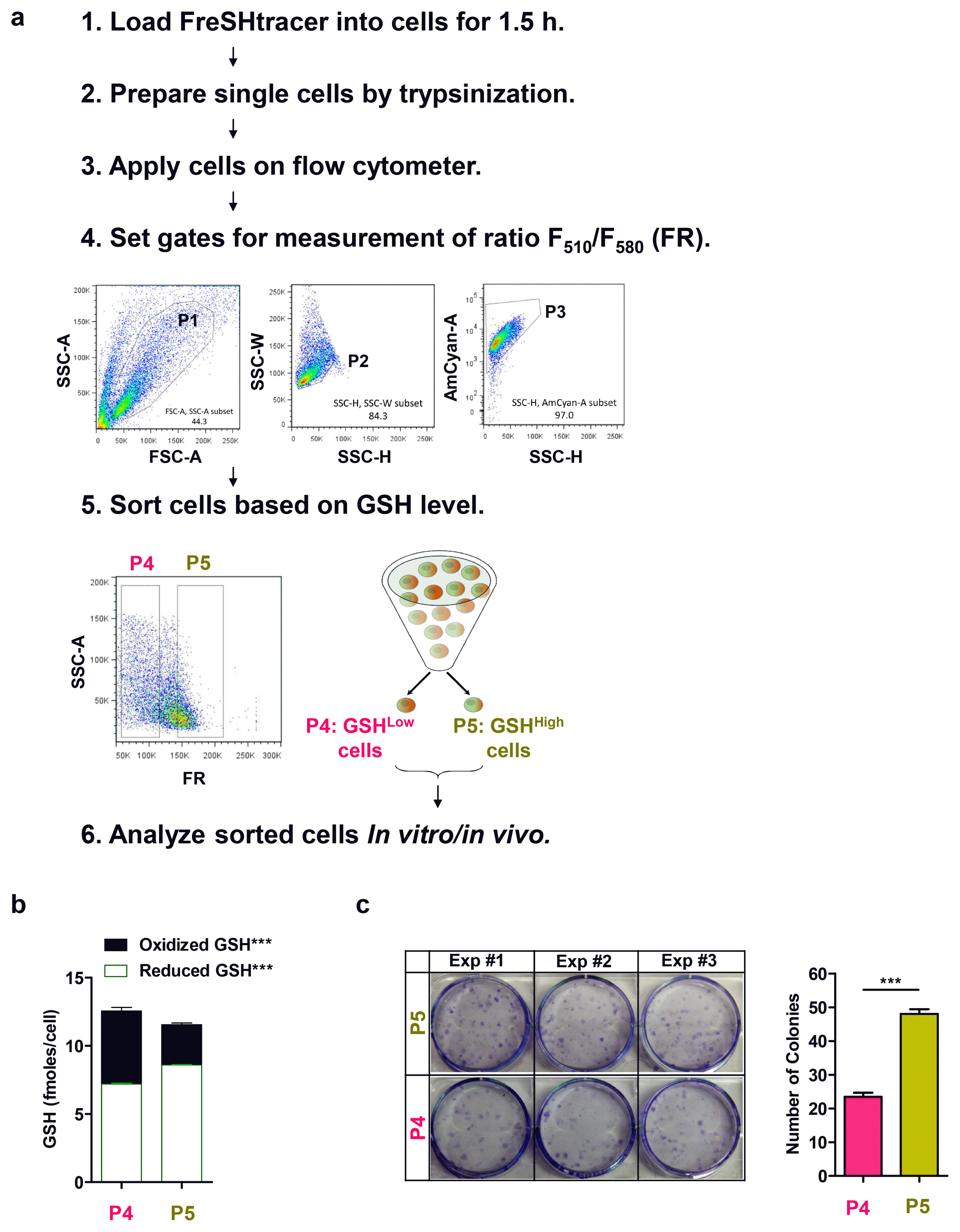
Comparison with other methods
Table 1
| Probe name | KD (mM, GSH) | Forward reaction rate constant (M−1s−1) | Quantum yield | Distribution in cells | Continuous monitoring time | Fluorophore | Publication year (reference) | |
|---|---|---|---|---|---|---|---|---|
|
|
||||||||
| GSH | PSH | |||||||
| Cyanoacrylamide-based probes | ||||||||
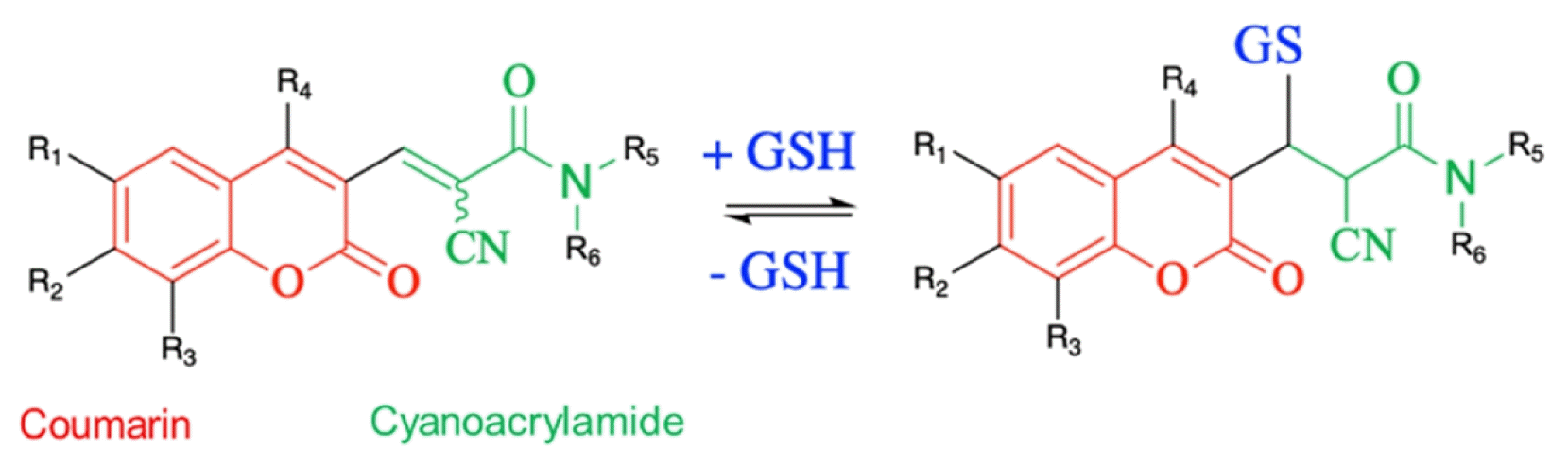
|
- Advantages: Can be targeted to subcellular organelles | |||||||
| - Disadvantages: Coumarin has been the only working fluorophore up to now | ||||||||
| FreSHtracer | 3.6 | 1.2 | 0.1 | 0.28 | Whole-cell | >several hours | Coumarin | 2012 (9), 2018 (10) |
| MitoFreSHtracer | 1.3 | N.D. | N.D. | N.D. | Mitochondria | <1 h | Coumarin | 2018 (10) |
| FreSHtracer-diAM | 3.8 | N.D. | N.D. | N.D. | Cytoplasm | <1 h | Coumarin | this paper |
| RealThiol | 3.7 | 7.5 | N.D. | 0.86 | Whole-cell | <15 min | Coumarin | 2017 (18) |
| Mito-RealThiol | 3.7 | Biphasic (102 → 0.82) | N.D. | 0.97 | Mitochondria | N.D. | Coumarin | 2017 (17) |
| HaloRT | 121 | N.D. | N.D. | N.D. | Various | N.D. | Coumarin | 2018 (25) |
|
|
||||||||
| α,β-unsaturated ketone-based probe | ||||||||
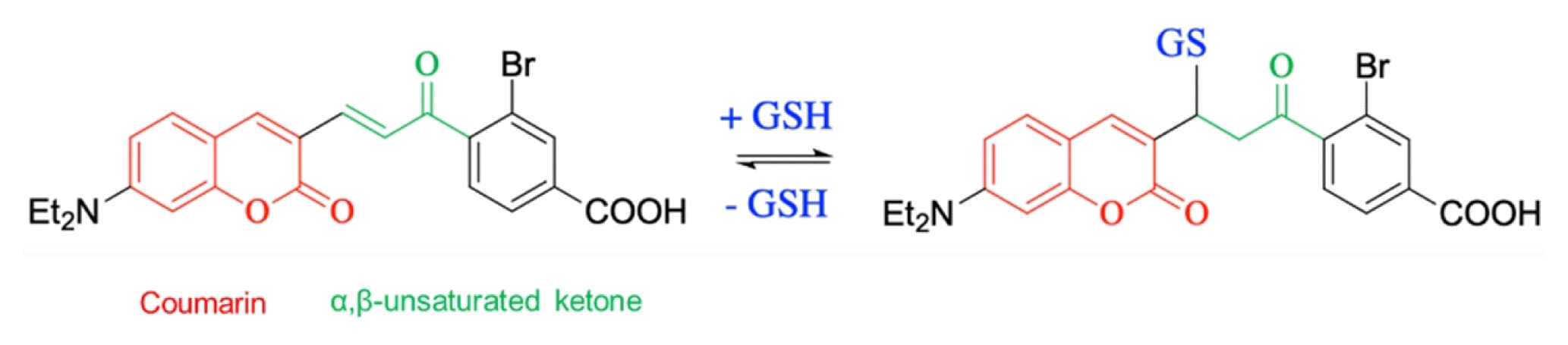
|
- Advantages: - | |||||||
| - Disadvantages: Slow reaction, low quantum yield | ||||||||
| TQ-Green | 1.6 | 0.15 | N.D. | 0.0059 | Whole-cell | N.D. | Coumarin | 2015 (19) |
|
|
||||||||
| Diarylcarbenium ion-based probe | ||||||||
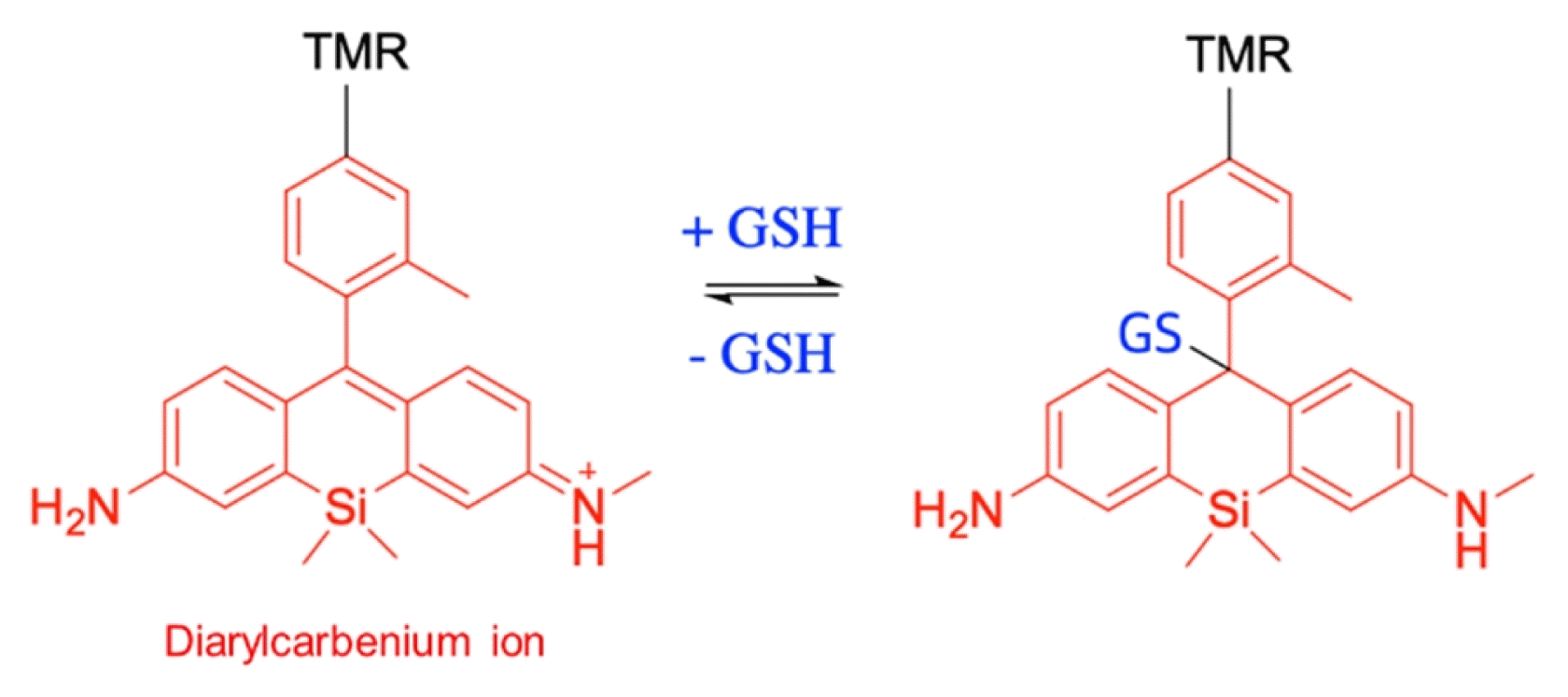
|
- Advantages: Fast reaction | |||||||
| - Disadvantages: Reports only mitochondrial GSH | ||||||||
| QG3.0 | 3.0 | 560 | N.D. | 0.27 | Mitochondria | N.D. | Rhodamine | 2017 (20) |
Experimental design and critical parameters
Limitations of the method
Materials and Methods
REAGENTS
DMSO (Merck Millipore, cat. no. 1.02931.1000)
FreSHtracer (Cell2in, cat. no. F-402)
MitoFreSHtracer (Cell2in, cat. no. M-377)
Cells of interest: in this study, we used hUC-MSCs from umbilical cord collected at the Obstetrics Department of Seoul National University Hospital (SNUH). The study was approved by the Internal Review Board (IRB) of the SNUH (IRB No. C-1708-083-878). CAUTION: Cells should be regularly checked to ensure that they are not infected with Mycoplasma.
Water, Ultra-Pure (Biosesang, cat. no. W2006)
MEM Alpha (Gibco, cat. no. 12561056)
FBS (Gibco, cat. no. 16000044)
Penicillin/streptomycin, 100× (Gibco, cat. no. 15140122)
HBSS (Welgene, cat. no. LB 003-02)
Dulbecco’s PBS×1 sterile solution (DPBS, pH 7.4; Welgene, cat. no. LB 001-02)
TrypLE™ Express Enzyme, 1×, no phenol red (Gibco, cat. no. 12604-013)
Trypan blue (Sigma-Aldrich, cat. no. T8154)
Dithiothreitol (DTT; Sigma-Aldrich, cat no. D9779)
Diamide (DA; Sigma-Aldrich, cat no. D3648)
N-Ethylmaleimide (NEM; Sigma-Aldrich, cat no. E1271)
Pluronic™ F-127 (Invitrogen, cat no. P6866)
HEPES (Corning, cat no. 25-060-Cl)
GSH-Glo™ Glutathione Assay kit (Promega, cat no. V6911)
Crystal Violet (Sigma, cat no. C3886)
EQUIPMENT
Confocal microscope (A1, Nikon)
Perfect Focusing System (PFS, Nikon)
Stage-top incubation system (Chamlide TC, Live Cell Instrument)
Laser sets for microscope imaging (Nikon; 405 and 488 nm)
Cover glass bottom dish (35 mm, SPL, cat. no. 200350)
Special glass cover for 35 mm culture dish (Chamlide TC, Live Cell Instrument, cat no. SG-C-10)
Clean bench (Labconco, class II type A2)
Centrifuge (Eppendorf, 5415R)
Centrifuge (Tomy, Flex-Spin LC-200)
Centrifuge tube (15 ml; SPL, cat no. 50015)
Centrifuge tube (50 ml; SPL, cat no. 50050)
Cell culture dish (100 mm; Corning, cat no. 430167)
Cell culture dish (150 mm; Sarstedt, cat no. 83.3903)
Six-well cell culture plate (Costar, cat no. 3516)
96-well plates (Corning white flat bottom; Sigma, cat no. CLS3922)
Pipet-Aid XP (Drummond Scientific Co, cat. no. 4-000-201-E)
Serological pipette (SPL, cat no. 91005, 91010, 91025)
Micropipettes (Thermo Scientific, cat no. 4641060, 4641080, 4641100)
Micropipette tips (Sorenson, cat no. 15730, 10040)
Hemocytometer (Marienfeld, cat. no. 0610039094)
FACS tube (5 ml Polystyrene Round-Bottom Tube 12×75 mm style; Falcon, cat. no. 352054)
FACS filter tube (5 ml Polystyrene Round-Bottom Tube with Cell-Strainer Cap; Falcon, cat. no. 352235)
Flow cytometer (BD FACS LSRFortessa)
Flow cytometer (BD FACS AriaIII)
Luminometer (TECAN Infinite M200 Pro)
REAGENT SETUP
EQUIPMENT SETUP
High-performance fluorescence microscopy system
Software for dual-excitation ratio image analysis
Flow cytometry
Software for flow cytometry data analysis
Characterization of sorted hUC-MSCs in vitro
PROCEDURE
Imaging and monitoring of GSH concentration in single living SCs
-
● TIMING: 2~3 h after seeding cells.
1│ Plate cells of interest on 35 mm cover glass bottom dishes. -
▲ CRITICAL STEP: Adjustment of the cell seeding density is crucial because cellular GSH concentration is sensitive to cell confluence (10).
2│ Load FreSHtracer (option A) or MitoFreSHtracer (option B) into cells as follows. ▲ CRITICAL STEP: FreSHtracers react not only with GSH but also with PSH. Whereas FreSHtracers equilibrate with GSH within 5 min, they show 8~12-fold slower reaction kinetics with PSH. Therefore, cells should be incubated with FreSHtracers for ≥1.0~1.5 h to equilibrate the probes with all the cellular thiols before measurements are made (10).
▲ CRITICAL STEP: If a CO2 supply is not available, cell culture media containing 10~25 mM HEPES could be used to minimize pH changes during the experiment. However, this alternative method is not recommended for the continuous monitoring of live cells for a long period of time.
-
? TROUBLESHOOTING
(A) Change the culture medium to 2 ml fresh medium containing 5 μM FreSHtracer and incubate for 1.5 h at 37°C in 5% CO2. -
▲ CRITICAL STEP: After this FreSHtracer-loading step, avoid washing the cells with buffers or medium that does not contain FreSHtracer.
(B) (i) Change the culture medium to fresh medium containing 10 μM MitoFreSHtracer and incubate for 1.5 h at 37°C, 5% CO2. ▲ CRITICAL STEP: Adding small amounts of Pluronic™ F-127, a non-ionic surfactant known to facilitate the solubilization of organic dyes, to the culture medium is helpful, but not required, for the staining of cells with MitoFreSHtracer.
▲ CRITICAL STEP: MitoFreSHtracer might also stain nuclei as the incubation time increases (>1 h).
-
? TROUBLESHOOTING
(ii) Discard the medium, wash the cells twice with HBSS, and then add 2 ml of fresh medium containing no MitoFreSHtracer.3│ Using a stage-top incubator, maintain the cells at 37°C in a humidified atmosphere containing 5% CO2 for 10~20 min, then visualize the cells using a confocal fluorescence microscope (for microscopy settings, see EQUIPMENT SETUP). ▲ CRITICAL STEP: Turn on the confocal microscope and lasers 1 h before image acquisition, and adjust the temperature to 37°C and the atmospheric CO2 content to 5%.
-
▲ CRITICAL STEP: Cellular GSH concentration is affected by thermal stress. Thus, it is important to maintain a constant temperature during the observation period. To prevent rapid changes in temperature, the volume of culture medium present in a 35 mm imaging dish should be at least 2 ml.
4│ Turn on the perfect focus system and select 3~4 cells. Stabilize the two-channel fluorescence intensities for 5 min at intervals of 5~30 s. ▲ CRITICAL STEP: Phototoxicity is an important concern for fluorescence microscopy experiments conducted in living cells. Therefore, adjustments of laser strength and time interval are required to avoid this.
-
? TROUBLESHOOTING
5│ Start image acquisition. -
? TROUBLESHOOTING
6│(Optional) Gently add a stock solution of the test compound (such as DA or DTT) into the culture medium (Fig. 2b and 2c). ▲ CRITICAL STEP: Large changes in the volume of media present can lead to focus drift. Therefore, the volume of added solution should not exceed 1% (20 μl in 2 ml of media) of that of the culture medium.
-
▲ CRITICAL STEP: The concentrations of the cell-permeable FreSHtracer in the medium and intracellularly are lowered by adding a stock solution of the test compound to the media. To prevent this dilution effect, the probe could be added to the stock solution at the same concentration as it is present in the culture medium. However, highly reactive test compounds may react with the probe, especially when present at high concentrations. Therefore, it is recommended that the stability of the probe is checked first (for the example of H2O2, see Supplementary Fig. S4) and the appropriate concentration of stock solution to be used is determined. By contrast, the MitoFreSHtracer signal is not affected by the addition of a test solution, because it is designed to be anchored inside the organelle.
7│ Analyze the acquired images using image-processing software, such as NIS-Elements or ImageJ.
Analysis of the GSH concentration in living SCs using flow cytometry
-
● TIMING: 3 h after seeding cells.
8│ Seed 5×104 cells in each well of a 6-well plate, add 3 ml of culture medium to each, and incubate overnight at 37°C in 5% CO2. -
▲ CRITICAL STEP: Prepare unstained cells simultaneously, which will be used to adjust flow cytomter parameters.
9│ Stain cells with FreSHtracer (option A) or Mito-FreSHtracer (option B) as follows. ▲ CRITICAL STEP: Turn on the flow cytometer 30 min before analyzing the samples to stabilize its optical system.
-
▲ CRITICAL STEP: FreSHtracers react not only with GSH but also PSH. Whereas FreSHtracers equilibrate with GSH within 5 min, they show 8~12-fold slower reaction kinetics with PSH. Therefore, cells should be incubated with FreSHtracers for ≥1.0~1.5 h to equilibrate the probes with all the cellular thiols before measurements are made (10).
(A) (i) Change the culture medium to 2 ml of fresh medium containing 2 μM FreSHtracer and incubate for 1.5 h at 37°C in 5% CO2.(ii) Wash cells with 3 ml of DPBS twice. Add 300 μl of TrypLE to each well and incubate at 37°C in 5% CO2 for 1~2 min until all the cells detach from the culture plate.(iii) Resuspend the cell pellet in 300 μl of DPBS containing 2% FBS and 4 μM FreSHtracer per well, then transfer the suspension to a FACS filter tube.(B) (i) Change the culture medium to 2 ml of fresh medium containing 5 μM MitoFreSHtracer and incubate for 1.5 h at 37°C in 5% CO2. -
? TROUBLESHOOTING
(ii) Wash the cells with 3 ml of DPBS twice. Add 300 μl of TrypLE to each well and incubate at 37°C in 5% CO2 for 1~2 min until all the cells detach from the culture plate.(iii) Resuspend the cell pellet in 300 μl of DPBS containing 2% FBS per well, then transfer to a FACS filter tube.10│ (Optional) Treat cells with ultra-pure water, 0.5 mM DA, or 0.5 mM DA and 0.5 mM DTT at the time points indicated in Fig. 3b, and incubate at room temperature (25°C).11│Keep cells on ice until flow cytometer analysis. PAUSE POINT: Keep cells at 4°C for a maximum of 30 min until they are loaded into the flow cytometer.
-
? TROUBLESHOOTING
12│ Adjust the flow cytometer settings as follows. Select FSC, SSC, AmCyan (F510), and PE (F580) parameters in the inspection window of the BD FACSDiva software. Vortex the unstained sample, load it into the sample injection port (SIP), and use it to optimize the voltages required for the application. Then, adjust the P1, P2, and P3 gates, and create a histogram for the FR value, as shown in Fig. 3a.13│ Analyze the stained samples using the optimized flow cytometer settings. ▲ CRITICAL STEP: The fluorescence intensities of FreSHtracers can be affected by temperature changes. Therefore, it is important to maintain a constant temperature (4°C) during flow cytometer analysis.
-
? TROUBLESHOOTING
14│ Export the raw data into FlowJo software in FCS format for further analysis.
Sort cells according to their GSH concentration using flow cytometry
-
● TIMING: 3~4.5 h after seeding the cells.
15│Seed cells into 150 mm dishes containing 25 ml of culture medium at a density of 30~60%. ▲ CRITICAL STEP: The typical yield of FreSHtracer-based cell sorting is <30%. Therefore, to obtain 1×106 cells from the upper and lower 30% of the entire cell population, 1~2×107 cells need to be sorted.
-
▲ CRITICAL STEP: Prepare unstained cells in one well of a 6-well plate.
16│ Change the culture medium to 25 ml of fresh medium containing 2 μM FreSHtracer and incubate for 1.5 h at 37°C in 5% CO2.17│ Turn on the cytometer 30 min before use to stabilize the system.18│ Install a 100 μm nozzle and stabilize the stream.19│ Set the drop delay using AccuDrop beads to assign the most appropriate value for sorting.20│ Adjust the plate voltages to locate the stream in the center of the collection tube.21│ Turn on the cooling and aerosol management systems for aseptic sorting at 4°C. -
▲ CRITICAL STEP: It takes about 45 min to cool the sample injection chamber.
22│ Wash cells with 25 ml (3 ml for the unstained sample) of DPBS twice. Add 3 ml (300 μl for the unstained cells) of TrypLE to a 150 mm dish (one well of a 6-well plate for the unstained sample) and incubate at 37°C in 5% CO2 for 1~2 min until all the cells detach after tapping the culture dish. Resuspend the cell pellet in 30 ml (3 ml for the unstained sample) of ice-cold culture medium containing 2.2 μM FreSHtracer (no probe for the unstained sample). Collect cells in 50 ml (15 ml for the unstained sample) tubes and count the cell number using a hemocytometer.23│ Centrifuge the cells at 500×g at 4°C for 5 min. -
▲ CRITICAL STEP: The centrifugal force and time that should be used are cell type-dependent.
24│ Discard the medium and resuspend the cell pellet in 1 ml of PBS containing 2% FBS and 2 μM FreSHtracer, and then transfer the suspension to a FACS filter tube.25│ Keep the tubes on ice until flow cytometer analysis.26│ Adjust the flow cytometer settings using the following steps: FSC-A versus SSC-A (P1) to capture all cells; SSC-A versus SSC-H (P2) to capture single cells; SSC-H versus AmCyan (P3) to capture all live cells; and SSC-A versus FR to capture cells according to their GSH content (P4, P5, etc.). Optimize the voltages for the application using the unstained sample. Examples of typical flow cytomter plots are shown in Fig. 4a.27│ Sort the cells on the basis of their FR value into 1 ml of culture medium in collection tubes. ▲ CRITICAL STEP: Fluorescence intensities of FreSHtracer can be affected by temperature changes. Therefore, it is important to maintain a constant temperature (4°C) during flow cytometer analysis.
-
? TROUBLESHOOTING
28│ For further functional studies, wash the sorted cells twice with ice-cold DPBS to remove FreSHtracer from the cells.29│ (Optional) To confirm the GSH concentration in the collected cells, analyze the lysates using an in vitro GSH assay kit, as described in EXPERIMENTAL SETUP (Fig. 4b).30│ (Optional) To test the in vitro function of the collected cells, analyze their CFU-F, as described in EXPERIMENTAL SETUP (Fig. 4c). -
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 2.Table 2
Troubleshooting table ● TIMING




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download