Abstract
Background and Objectives
This study aims to explore the effects of a long non-coding RNA, LINC00525, on colorectal cancer (CRC) and its underlying molecular mechanisms.
Methods
The qPCR, MTT, colony formation, Western blotting, Luciferase reporter and biotin pull-down, shRNA knockdown and DNA fragmentation assays were performed in this study.
Results
High expressions of LINC00525 were associated with poor prognosis of CRC patients. LINC00525 knockdown decreased stemness properties and increased sensitivities to oxaliplatin. MiR-507 was a direct target of LINC00525 and overexpression of miR-507 significantly decreased abilities of tumorsphere formation and cell growth. Overexpression of miR-507 resulted in a decrease of expression of cancer stem cell markers and the increase of apoptosis rates. MiR-507 regulated the expression of ELK3. In addition, LINC00525 knockdown decreased the expression of ELK3. Restoration of ELK3 expression abrogated the effects of LINC00525 knockdown. LINC00525 could be served as prognostic marker of CRC.
Most of the colorectal cancer (CRC)-related deaths are caused by recurrence and metastasis of CRC and its chemotherapy resistance (1). Oxaliplatin is a third-generation platinum compound widely used for the treatment of CRC in clinical studies (2). When oxaliplatin combined with 5-flourouracil and folinic acid for the treatment of metastatic CRC, the median progression-free survival of CRC patients prolonged 2 years (3). Despite this improvement, all metastatic CRC finally developed to oxaliplatin resistance and patients succumbed to their disease (4). Therefore, identification of the underlying molecular mechanisms of CRC chemotherapy resistance is worthwhile for its therapy and prevention strategies.
Cancer stem cells (CSCs) have been identified to play a crucial role in a variety of cancers including breast cancer, ovarian, melanoma and CRC (5, 6). CSCs are naturally resistant to chemotherapy due to their ability for DNA repair and multidrug resistance ABC transporters expression (7). Those CSCs confer drug resistance to their progeny, resulting in the tumors eventually become less sensitive to the following chemotherapy (7, 8). An important strategy to overcome drug resistance is to regulate CSCs stemness and thereby to increase the sensitivities of the tumor to chemotherapy.
It has been identified that long non-coding RNA (lncRNA) expression pattern is highly correlated with cancer development and prognosis (9, 10). More recently, some studies have revealed that lncRNAs are capable of regulation of CSCs stemness via different molecular mechanisms (11, 12). It is known that CSCs stemness is a major contributor to cancer chemotherapy resistance (7). Therefore, identification of novel lncRNAs which function as the regulators for CSCs stemness might provide evidence for the development of strategies to overcome cancer chemotherapy resistance. Additionally, abnormal expressions of LINC00525 have been identified in the development of many types of cancer including bladder cancer (13), lung cancer (14) and leukemia (15). However, the roles of LINC00525 in the occurrence and development of CRC are still unknown. Thus, in this study, for the first time, we investigated the relationships between LINC00525 and CRC.
Fresh CRC specimens and normal intestine tissue samples were collected from Yidu Central Hospital of Weifang City between 2017 to 2018. In addition, in Fresh CRC specimens, twenty-eight samples were collected from oxaliplatin sensitive patients and thirty-two samples were collected from oxaliplatin resistant patients. The study was approved by the ethics commitment of Yidu Central Hospital of Weifang City, and followed the ethical guidelines for medical research on Human subjects, the Helsinki Declaration of 1975, as revised in 2000.
Five human CRC cell lines including Caco2, SW480, SW620, HCT116 and HT29 and normal human intestinal epithelial cell line (HIEC) were purchased from the Cell Bank of the Chinese Academic of Sciences (Shanghai, China). The cells were cultured in recommended medium supplemented with 10% fetal bovine serum (FBS) (Life Technologies, Gaithersburg, MD, USA) and 100 U/ml penicillin and 100 μg/ml streptomycin (Beyotime, Shanghai, PR China) at 37°C in the presence of 5% CO2 at constant humidity.
Tumorsphere medium used in this study consists of Dulbecco’s Modified Eagle Medium (DMEM)/F12, epidermal growth factor (20 ng/ml), insulin (5 μg/ml), 2% B-27 and 0.4% FBS. Tumorsphere formation assay was performed according to the previously reported method (16). In brief, the HCT116 and HT29 cells were harvested and suspended in phosphate-buffered saline (PBS). Next, dissociated cells were then seeded in tumorsphere medium. The spheres were observed under an inverted microscope.
MTT and colony formation were performed according to previously reported methods (17, 18). In brief, MTT solution was added into each well and the microplate was incubated at 37°C for 4 h. DMSO solution was then added and the optical density was read at 570 nm using a microplate reader (Molecular Devices, Sunnyvale, CA, United States). For colony formation assay, HCT116 and HT29 cells were seeded in 6-well plate and grown for indicated days. Next, the colonies were fixed with 4% paraformaldehyde and then stained with crystal violet (0.05% w/v) overnight. The numbers of colonies were counted using Image J.
DNA fragmentation ELISA kit (Roche, Indianapolis, IN, United States) was used to evaluate cell apoptosis, according to the manufacturer’s instructions.
To construct LINC00525 knockdown cell lines, the cells were transfected with lentiviruses expressing either LINC00525-1 shRNA or LINC00525-2 shRNA. The shRNA sequences are in Supplementary Table S1. Twenty-four hours after transfection, total RNA was isolated and expressions of LINC00525 were analyzed by qPCR.
To construct LINC00525 and ELK3 overexpression cell lines, LINC00525 or ELK3 was amplified and subcloned into p-Sin 3×flag vector. The cells were transfected with p-Sin 3×flag vector containing LINC00525 or ELK3 and followed by the selection of antibiotics. Primers for amplification of LINC00525 and ELK3 are shown in Supplementary Table S1. Expressions of LINC00525 and ELK3 were determined by either qPCR or western blotting, to ensure the stable cell lines were successfully established.
Biotin pull-down assay was performed under RNase free conditions according to previously reported methods (19). Sense and anti-sense biotin-labeled DNA oligomers were used. Three pairs of LINC00525-DNA oligomers were used in this study. The sequences are shown in Supplementary Table S1. Biotin-labeled DNA oligomers were incubated with cell lysates for 1 h. Next, agarose beads conjugated with streptavidin were added to isolate RNA-RNA complex.
Dual-luciferase® reporter assay system was established according to the manufacturer’s instructions (Promega, Madison, MA, United States). LINC00525 wild-type (LINC00525 wt) and the miR-507-binding-site mutated (LINC00525 mt) one was constructed and then integrated into dual luciferase vector. After that, the cells were co-transfected with dual luciferase vector and miR-507 or miR-NC mimics.
Similarly, dual luciferase vector containing wild-type ELK3 3′ UTR or mutant ELK3 3′ UTR was established. Next, the cells were co-transfected with dual luciferase vector and miR-507 or miR-NC mimics. The relative luciferase activities were detected after 48 h.
Trizol reagent was used to extract total RNA from clinical specimens and cell lines, according to the manufacturer’s instructions (Takara, Japan). To remove DNA contamination, RNase-free DNase I was used. Primers for LINC00525, ELK3, CD44, SOX2, OCT4, miR-507, and internal control including GADPH and U6 were used for amplification of these genes. The sequences are presented in Supplementary Table S1. To analyze the accuracy of the PCR reaction, the Melt curves were used. To evaluate the expressions of genes, 2−ΔΔCt values were calculated. The mRNA expression values of LINC00525, CD44, SOX2, and OCT4 were normalized to that of GADPH. The mRNA expression value of miR-507 was normalized to that of U6.
Extraction and qualification of proteins were performed according to the previous methods (20). After the membrane was probed with antibodies against LINC00525, CD44, SOX2, OCT4, ELK3, p53, Caspase-3, Bcl-2, and actin overnight at 4°C, the membrane was then incubated with appropriated secondary antibodies. ChemiDoc MP imaging system (Bio-Rad, Hercules, CA, USA) was used to examine chemiluminescence.
To our knowledge, the roles of LINC00525 in the occurrence and development of CRC are still unknown. Thus, we first explored the relationships between LINC00525 expressions and prognosis of CRC. The relative expressions of LINC00525 were determined in colorectal tumor specimen (n=123) and matched healthy tissues (n=25) from GSE21510 dataset. Expressions of LINC00525 in CRC were significantly increased when compared with that in the healthy tissues (Fig. 1a, p<0.001). In addition, the relative expressions of LINC00525 in colorectal tumor specimen (n=60) and normal intestinal tissues (n=60) were investigated. The expressions of LINC00525 in CRC were significantly increased when compared with that in the normal intestinal tissues (Fig. 1b, p<0.01). We investigated LINC00525 expressions in CRC tissues of oxaliplatin sensitive (OxS) patients (n=32) and oxaliplatin resistant (OxR) patients (n=28). Interestingly, the expressions of LINC00525 in the CRC tissues from OxR patients were higher than those in the CRC tissues from OxS patients (Fig. 1c).
The results further demonstrated that LINC00525 expression levels were higher in CRC cell lines than those in the HIEC (Fig. 1d). We further explored the LINC00525 expression pattern in sphere-forming HTC116 and HT29 cells and their parent cells. Interestingly, LINC00525 expression levels were significantly increased in sphere-forming HTC116 and HT29 cells when compared with those in the parent cells (Fig. 1e, p<0.01).
Furthermore, Kaplan-Meier’s analysis demonstrated that high expressions of LINC00525 were associated with a low survival rate, whereas the low expressions of LINC were associated with high survival rate (Fig. 1f, p=0.0154). Taken together, these results supported that LINC00525 served as a marker for poor prognosis of CRC and could be involved in the regulation of CRC stemness and oxaliplatin resistance.
First, as shown in Supplementary Fig. S1, we successfully constructed LINC00525 overexpressed HCT116 and HT29 cells (Supplementary Fig. S1a and S1b). Interestingly, the results demonstrated that the mRNA and protein expressions of cancer stem cell markers including CD44, SOX2 and OCT4 were significantly increased in LINC00525 overexpressed HCT116 and HT29 cells when compared with those in the control group (Supplementary Fig. S1c~e). These results supported that that overexpression of LINC00525 was associated with the stemness of colorectal cancer.
We then knocked down LINC00525 in HCT116 and HT29 cells. The results showed that mRNA expressions of LINC00525 in LINC00525 shRNA-transfected cells were significantly decreased when compared with those in the control shRNA-transfected cells, indicating that LINC00525 was successfully knocked down in these cells (Fig. 2a, p<0.01).
The results also demonstrated that LINC00525 knockdown significantly decreased the capacity to form tumorsphere (Fig. 2b, p<0.01) when compared with those in the control group. In addition, the results showed that expressions of cancer stem cell markers including CD44, SOX2, and OCT4 were significantly decreased at both the transcriptional (Fig. 2c and 2d, p<0.01) and post-transcriptional levels (Fig. 2e, p<0.01) in LINC00525 knockdown cells when compared with those in the control group. However, downregulation of protein expressions of SOX2, and OCT4 might be due to the decrease of SOX2, and OCT4 mRNA. Therefore, the regulatory effects of LINC00525 on the protein expressions of SOX2 and OCT4 are directly or indirectly should be clarified in further study.
The results showed that mRNA expressions of LINC00525 in LINC00525 shRNA-transfected cells including HCT116-OxR and HT29-OxR cells were significantly decreased when compared with those in the control shRNA-transfected cells, indicating that LINC00525 was successfully knocked down in these cells (Fig. 2f, p<0.01).
Next, the results demonstrated that LINC00525 knockdown significantly decreased cell viabilities and colony formation abilities of OxR-HCT116 and OxR-HT29 cells when compared with those of the OxR-HCT116 and OxR-HT29 cells transfected with control shRNA (Fig. 2g~ i, p<0.01). LINC00525 knockdown also significantly increased apoptotic cell populations in OxR-HCT116 and OxR-HT29 cells (Fig. 2j, p<0.01). In addition, LINC00525 knockdown increased the expressions of p53 and Caspase-3, decreased the expressions of Bcl-2 when compared with those of the OxR-HCT116 and OxR-HT29 cells transfected with control shRNA (Fig. 2k).
All results suggested that LINC00525 knockdown inhibited the stemness and oxaliplatin resistance of CRC.
We screened the target miRNAs of LINC00525 using miRcode and identified miR-507 as a target of LINC00525. As shown in Fig. 3a, the binding sites between LINC00525 transcript and miR-507 were predicted using RNAhybrid. Dual luciferase reporter assay showed that miR-507 mimics dramatically decreased the luciferase activities of LINC00525 wt in the HCT116 (Fig. 3b, p< 0.01) and HT29 cells (Fig. 3c, p<0.01). However, luciferase activities between miR-507 mimics and miR-NC mimics in the cells transfected with LINC00525 mt showed no significant difference. These results supported that LINC00525 interacted with miR-507.
Next, we found that LINC00525 knockdown significantly increased the expressions of miR-507 in the HCT116 and HT29 cells (Fig. 3d, p<0.01). Interestingly, when LINC00525 was overexpressed in the cells, the expressions of miR-507 were significantly decreased (Fig. 3e, p<0.01). RNA-pulldown assay was performed to investigate whether LINC00525 directly interacted with miR-507. The results demonstrated that the relative expressions of LINC00525 and miR-507 were dramatically increased in anti-sense DNA probes group, indicating that LINC00525 directly interacted with miR-507 in the HCT116 and HT29 cells (Fig. 3f and 3g, p<0.01). Additionally, the relative expressions of LINC00525 in the HCT116 and HT29 cells were significantly decreased when the cells treated with miR-507 mimics (Fig. 3h).
CRC cells were transfected with miR-507 mimics or mimics negative control (miR-NC). Relative expressions of miR-507 were significantly increased in the cells transfected with miR-507 mimics when compared with that in cells transfected with miR-NC (Fig. 4a, p<0.001). Next, we found that overexpression of miR-507 significantly decreased the capacity to form tumorspheres in HCT116 and HT29 cells (Fig. 4b, p<0.01). Additionally, expressions of cancer stem cell markers were evaluated in the cells transfected with miR-507 mimics or miR-NC. The results demonstrated that overexpression of miR-507 significantly decreased the expressions of cancer stem cell markers including CD44, SOX2, and OCT4 at both the transcriptional (Fig. 4c and 4d, p<0.01) and post-transcriptional levels (Fig. 4e).
HCT116-OxR and HT29-OxR cells with miR-507 overexpression were successfully constructed (Fig. 4f). The results demonstrated that miR-507 overexpression significantly suppressed cell viabilities and colony formations when compared with that of miR-NC transfected cells (Fig. 4g~i, p<0.01). Additionally, miR-507 overexpression significantly increased cell apoptosis rates when compared with that of miR-NC transfected cells (Fig. 4j, p<0.01). miR-507 overexpression also increased the expressions of p53 and Caspase-3, decreased the expressions of Bcl-2 when compared with those of the OxR-HCT116 and OxR-HT29 cells transfected with miR-NC (Fig. 4k). Taken together, these data demonstrated that miR-507 inhibited the CRC stemness and oxaliplatin resistance in HCT116 and HT29 cells.
Targetscan was used to predict the possible targets of miR-507 and ELK3 was identified. The binding sites between miR-507 and ELK3 were shown in Fig. 5a. Additionally, the dual luciferase reporter assay demonstrated that miR-507 mimics specifically decreased the luciferase activities of wide-type ELK 3′-UTR in the HCT116 (Fig. 5b, p<0.01) and HT29 cells (Fig. 5c, p<0.05), indicating that effects of miR-507 on ELK3 expression.
The results demonstrated that ELK3 mRNA levels were significantly increased in bio-miR-507 pulldown group (Fig. 5d, p<0.05), indicating that miR-507 interacted with ELK3. In addition, overexpression of miR-507 significantly decreased the relative expressions of ELK (Fig. 5e, p<0.05). Interestingly, the results demonstrated that inhibition of miR-507 significantly decreased the relative expressions of ELK in the HCT116 (Fig. 5f, p<0.01) and HT29 cells (Fig. 5g, p<0.01).
We then investigated the effects of LINC00525 on the expression pattern of ELK3. The results demonstrated that LINC00525 knockdown significantly decreased the mRNA and protein expressions of ELK3 (Fig. 5h and 5j, p<0.01), whereas LINC00525 overexpression suppressed the mRNA expressions of ELK3 (Fig. 5i, p<0.01). The effects of LINC00525 on the regulation of ELK are at the transcriptional levels (Fig. 5h~i). However, since the decrease of mRNA of ELK leaded to the decrease of protein expression of ELK. Therefore, the regulatory effects of LINC00525 on the protein expressions of ELK should be clarified in further study.
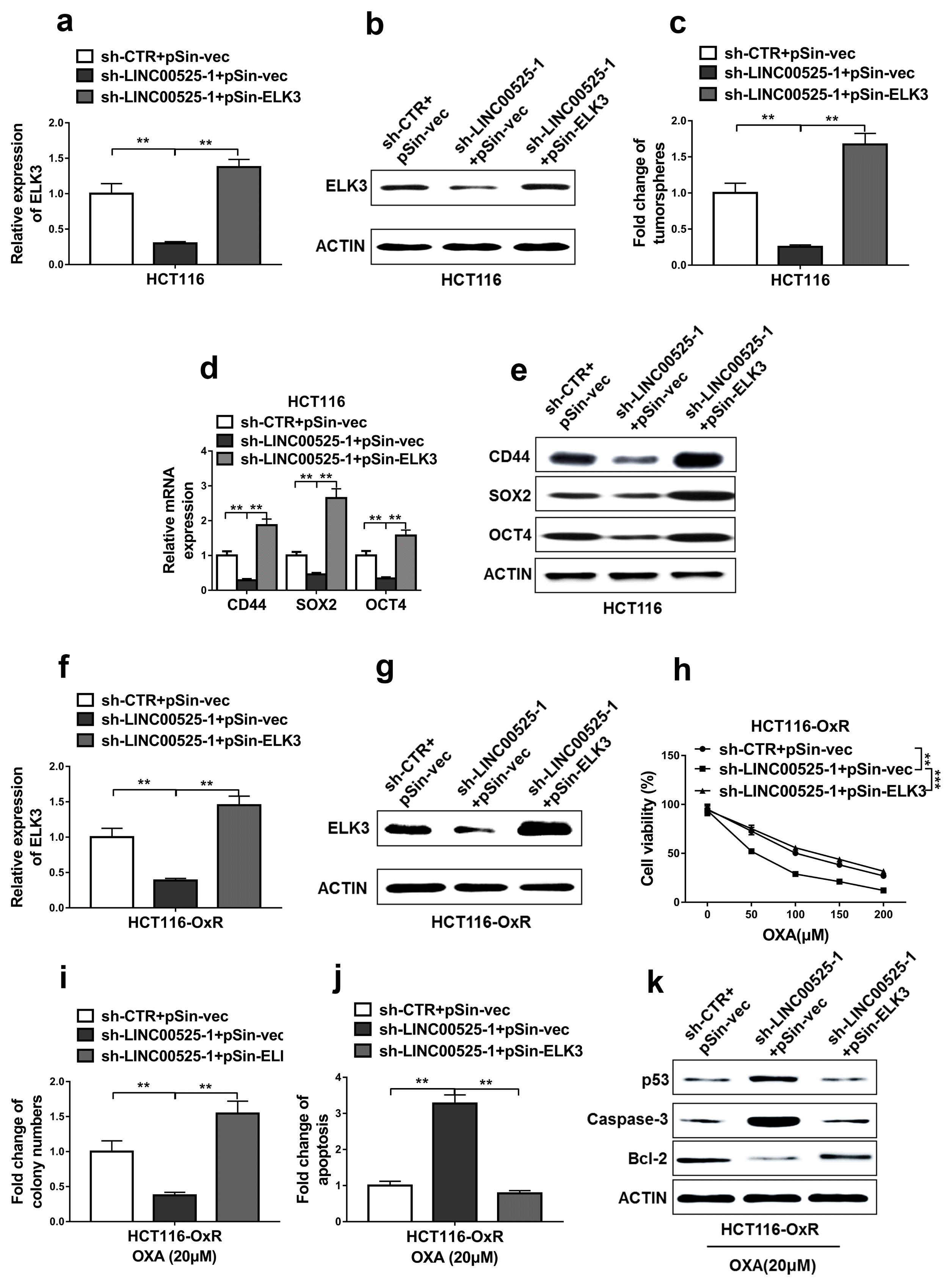
First, we successfully restored the ELK3 expression in the HCT116 cells transfected with LINC00525 shRNA (Fig. 6a and 6b). The results demonstrated that restoration of ELK3 expression significantly increased the capacity to form tumorsphere of LINC00525 knockdown cells (Fig. 6c). Additionally, restoration of ELK3 expression also significantly increased the expressions of cancer stem cell markers including CD44, SOX2, and OCT4 in the LINC00525 knockdown cells (Fig. 6d and 6e).
HCT116-OxR cells were successfully transfected with pSin-ELK3, as shown in Fig. 6f and 6g. Next, cell viabilities and apoptosis were evaluated. The results demonstrated that restoration of ELK3 expression increased the cell viabilities and colony formations in LINC00525 knockdown cells when compared with that of LINC00525 knockdown cells transfected with pSin-vec (Fig. 6h and 6i). Interestingly, restoration of ELK3 expression in HCT116-OxR cells transfected with LINC00525 shRNA significantly decreased the apoptosis rates when compared with HCT116-OxR cells cotransfected with LINC00525 shRNA and pSin-vec (Fig. 6j). Apart from that, restoration of ELK3 expression in HCT116-OxR cells transfected with LINC00525 shRNA also significantly decreased the expressions of p53 and Caspase-3, increased the expressions of Bcl-2 when compared with HCT116-OxR cells cotransfected with LINC00525 shRNA and pSin-vec (Fig. 6k). These results supported that restoration of ELK3 expression abrogated the effects of LINC00525 knockdown in oxaliplatin resistant CRC cells.
As the third-generation of platinum compound, oxaliplatin exhibits promising properties in the treatment of CRC (21). In clinical trials, oxaliplatin produces a response rate of 12% to 24% for patients with previously untreated CRC (22). When it combined with 5-flourouracil, more than 50% response rate of metastasis CRC patients is reported (23). Recently, strategies have also been developed to combine oxaliplatin with monoclonal antibodies including panitumumab, cetuximab, and bevacizumab in the treatment of metastasis CRC patients (24). Unfortunately, metastatic CRC patients finally developed to oxaliplatin resistance and succumbed to the disease (4). Therefore, identification of some regulators able to overcome oxaliplatin resistance can be used to improve the response rate of CRC patients.
CSCs are naturally resistant to chemotherapy due to their ability for DNA repair and multidrug resistance ABC transporters expression (7). Those CSCs confer drug resistance to their progeny, resulting in the tumors eventually become less sensitive to the following chemotherapy (7, 8). Therefore, strategies for regulation of CSC stemness can be applied to overcome oxaliplatin resistance. In the current study, we identified a novel lncRNA, LINC00525, as a regulator for CRC stemness. First, we found that expressions of LINC00525 in the CRC tissues from OxR patients were much higher than those in the CRC tissues from OxS patients. Interestingly, we observed that LINC00525 knockdown dramatically decreased cancer stem cell markers including CD44, SOX2, and OCT4 at both the transcriptional and post-transcriptional levels. More importantly, LINC00525 knockdown resulted in the decrease of cell viabilities and colony formation abilities, the increase of cell apoptosis rate in the oxaliplatin resistant cells. These results supported that LINC00525 was associated with CRC stemness. LINC00525 knockdown can be used to regulate CRC stemness, thereby increasing sensitivities of CRC to oxaliplatin.
One important function of lncRNAs is to act as endogenous miRNA sponges (25). LncRNA can regulate miRNA activities by lncRNA-miRNA interactions (26, 27). Therefore, in the current study, we explored the possible target miRNAs of LINC00525 using miRcode and identified miR-507 as a target of LINC00525. One study performed by Jia et al. (28) has demonstrated that miR-507 regulates cell migration and invasion in breast cancer cells. In another study, Wang et al. (29) have reported that miR-507 inhibits the levels of vascular endothelial growth factor C in human chondrosarcoma cells. The current study supported that miR-507 regulated the stemness and oxaliplatin resistance of CRC in part by ELK3 suppression. Our results, along with the previous studies, suggested that miR-507 functions an important marker for the development of different types of cancers. More importantly, we found that a novel lncRNA, LINC00525, regulated the expressions of miR-507, thereby regulating CRC stemness and oxaliplatin resistance.
ELK3 is a transcription factor belonging to Ets family (30). It can be activated by different signaling pathways including PI3K/Akt/mammalian target of rapamycin (mTOR) and ERK signaling pathways (31). Some studies have identified that ELK3 is involved in the incidence and progression of different types of cancers. For instance, it has reported that ELK3 regulates cell migration and invasion in breast cancer and hepatoma cells (32) The current study demonstrated that ELK3 was involved in the regulation of CRC stemness and resistance to oxaliplatin. We found that ELK3 was a direct target of miR-507 and restoration of its expression abrogated the effects of LINC00525 knockdown. Interestingly, another study performed by Park et al. (33) has reported that ELK3 suppression regulates sensitivities of breast cancer cells to doxorubicin. These results suggest that ELK plays an important role in the regulation of cancer cell chemotherapy resistance. More importantly, the results showed that miR-507 regulated expression of ELK3. Inhibition of miR-507 significantly increased the expressions of ELK3. Therefore, apart from PI3K/Akt/mTOR and ERK-mediated signaling pathways, we identified that the inhibition of miR-507 is another pathway triggering ELK3 activation.
This study demonstrated that LINC00525 functioned as an oncogene and its high expression was associated with prognosis of CRC patients. LINC00525 promoted the stemness and chemoresistance of CRC by targeting miR-507/ELK3 axis. Silencing LINC00525 inhibited the stemness and oxaliplatin resistance of CRC. These results supported that LINC00525 might serve as a prognostic biomarker and a potential target for CRC therapy.
Notes
Supplementary Materials
Supplementary data including one table and one figure can be found with this article online at http://pdf.medrang.co.kr/paper/pdf/IJSC/IJSC-12-s19041.pdf.
References
1. Midgley R, Kerr D. Immunotherapy for colorectal cancer: a challenge to clinical trial design. Lancet Oncol. 2000; 1:159–168. DOI: 10.1016/S1470-2045(00)00034-6. PMID: 11905654.

2. Meyerhardt JA, Mayer RJ. Systemic therapy for colorectal cancer. N Engl J Med. 2005; 352:476–487. DOI: 10.1056/NEJMra040958. PMID: 15689586.

3. Hwang IG, Jang JS, Oh SY, Rho MH, Lee S, Park YS, Park JO, Nam EM, Lee HR, Jun HJ, Chi KC. Phase II study of mFOLFOX3 (5-fluorouracil, leucovorin, oxaliplatin) as second-line treatment after gemcitabine failure in patients with unresectable/metastatic biliary tract cancer. Cancer Chemother Pharmacol. 2015; 75:757–762. DOI: 10.1007/s00280-015-2691-1. PMID: 25677446.

4. Chocry M, Leloup L, Kovacic H. Correction: reversion of resistance to oxaliplatin by inhibition of p38 MAPK in colorectal cancer cell lines: involvement of the calpain/Nox1 pathway. Oncotarget. 2018; 9:26978–26979. DOI: 10.18632/oncotarget.25605. PMID: 29928496. PMCID: PMC6003572.

5. Prager BC, Xie Q, Bao S, Rich JN. Cancer stem cells: the architects of the tumor ecosystem. Cell Stem Cell. 2019; 24:41–53. DOI: 10.1016/j.stem.2018.12.009. PMID: 30609398. PMCID: PMC6350931.

6. Al-Alem LF, Pandya UM, Baker AT, Bellio C, Zarrella BD, Clark J, DiGloria CM, Rueda BR. Ovarian cancer stem cells: what progress have we made? Int J Biochem Cell Biol. 2019; 107:92–103. DOI: 10.1016/j.biocel.2018.12.010. PMID: 30572025.

7. Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005; 5:275–284. DOI: 10.1038/nrc1590. PMID: 15803154.

8. Phi LTH, Sari IN, Yang YG, Lee SH, Jun N, Kim KS, Lee YK, Kwon HY. Cancer Stem Cells (CSCs) in drug resistance and their therapeutic implications in cancer treatment. Stem Cells Int. 2018; 2018:5416923. DOI: 10.1155/2018/5416923. PMID: 29681949. PMCID: PMC5850899.

9. Hon KW, Abu N, Ab Mutalib NS, Jamal R. miRNAs and lncRNAs as predictive biomarkers of response to FOLFOX therapy in colorectal cancer. Front Pharmacol. 2018; 9:846. DOI: 10.3389/fphar.2018.00846. PMID: 30127741. PMCID: PMC6088237.

10. Li CH, Chen Y. Diagnostic potential of lncRNAs in cancer. EBioMedicine. 2016; 7:7–8. DOI: 10.1016/j.ebiom.2016.05.013. PMID: 27322445. PMCID: PMC4909482.

11. Chen S, Zhu J, Wang F, Guan Z, Ge Y, Yang X, Cai J. LncRNAs and their role in cancer stem cells. Oncotarget. 2017; 8:110685–110692. DOI: 10.18632/oncotarget.22161. PMID: 29299179. PMCID: PMC5746414.

12. Castro-Oropeza R, Melendez-Zajgla J, Maldonado V, Vazquez-Santillan K. The emerging role of lncRNAs in the regulation of cancer stem cells. Cell Oncol (Dordr). 2018; 41:585–603. DOI: 10.1007/s13402-018-0406-4. PMID: 30218296.

13. Xu Z, Wang C, Xiang X, Li J, Huang J. Characterization of mRNA expression and endogenous RNA profiles in bladder cancer based on The Cancer Genome Atlas (TCGA) database. Med Sci Monit. 2019; 25:3041–3060. DOI: 10.12659/MSM.915487. PMID: 31020952. PMCID: PMC6498884.

14. Wang J, Zhang Y, Wan H, Dong N, Bao L, Sun X, Xu M, Wang X. Global analyses of subtype- or stage-specific genes on chromosome 7 in patients with lung cancer. Cancer Metastasis Rev. 2015; 34:333–345. DOI: 10.1007/s10555-015-9568-y. PMID: 25951983.

15. Labaj W, Papiez A, Polanska J, Planski A. Deep data analysis of a large microarray collection for leukemia biomarker identification. Paper presented at: 10th International Conference on Practical Applications of Computational Biology & Bioinformatics. 2016 Jun 1–3; Seville, Spain.

16. Liu L, Liu J, Wang H, Zhao H, Du Y. Fenretinide targeting of human colon cancer sphere cells through cell cycle regulation and stress-responsive activities. Oncol Lett. 2018; 16:5339–5348. PMID: 30250604. PMCID: PMC6144862.

17. Yang H, Liu C, Zhang YQ, Ge LT, Chen J, Jia XQ, Gu RX, Sun Y, Sun WD. Ilexgenin A induces B16–F10 melanoma cell G1/S arrest in vitro and reduces tumor growth in vivo. Int Immunopharmacol. 2015; 24:423–431. DOI: 10.1016/j.intimp.2014.12.040. PMID: 25596038.

18. Xu K, Chen G, Qiu Y, Yuan Z, Li H, Yuan X, Sun J, Xu J, Liang X, Yin P. miR-503-5p confers drug resistance by targeting PUMA in colorectal carcinoma. Oncotarget. 2017; 8:21719–21732. PMID: 28423513. PMCID: PMC5400618.

19. Yamamoto K, Ito S, Hanafusa H, Shimizu K, Ouchida M. Uncovering direct targets of MiR-19a involved in lung cancer progression. PLoS One. 2015; 10:e0137887. DOI: 10.1371/journal.pone.0137887. PMID: 26367773. PMCID: PMC4569347.

20. Shang W, Jiang Y, Boettcher M, Ding K, Mollenauer M, Liu Z, Wen X, Liu C, Hao P, Zhao S, McManus MT, Wei L, Weiss A, Wang H. Genome-wide CRISPR screen identifies FAM49B as a key regulator of actin dynamics and T cell activation. Proc Natl Acad Sci U S A. 2018; 115:E4051–E4060. DOI: 10.1073/pnas.1801340115. PMID: 29632189. PMCID: PMC5924929.

21. Bruera G, Ricevuto E. Intensive chemotherapy of metastatic colorectal cancer: weighing between safety and clinical efficacy: evaluation of Masi G, Loupakis F, Salvatore L, et al. Bevacizumab with FOLFOXIRI (irinotecan, oxaliplatin, fluorouracil, and folinate) as first-line treatment for metastatic colorectal cancer: a phase 2 trial. Lancet Oncol. 2011; 11:821–824. DOI: 10.1517/14712598.2011.582462. PMID: 21545334.

22. Becouarn Y, Rougier P. Clinical efficacy of oxaliplatin monotherapy: phase II trials in advanced colorectal cancer. Semin Oncol. 1998; 25(2 Suppl 5):23–31. PMID: 9609105.
23. deBraud F, Munzone E, Nolè F, De Pas T, Biffi R, Brienza S, Aapro MS. Synergistic activity of oxaliplatin and 5-fluorouracil in patients with metastatic colorectal cancer with progressive disease while on or after 5-fluorouracil. Am J Clin Oncol. 1998; 21:279–283. DOI: 10.1097/00000421-199806000-00015. PMID: 9626798.

24. Comella P, Casaretti R, Sandomenico C, Avallone A, Franco L. Role of oxaliplatin in the treatment of colorectal cancer. Ther Clin Risk Manag. 2009; 5:229–238. DOI: 10.2147/TCRM.S3583. PMID: 19436599. PMCID: PMC2697505.

25. Tian L, Hu X, He Y, Wu Z, Li D, Zhang H. Construction of lncRNA-miRNA-mRNA networks reveals functional lncRNAs in abdominal aortic aneurysm. Exp Ther Med. 2018; 16:3978–3986. PMID: 30344676. PMCID: PMC6176170.

26. Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA-LncRNA interactions. Methods Mol Biol. 2016; 1402:271–286. DOI: 10.1007/978-1-4939-3378-5_21. PMID: 26721498.

27. Guo H, Qi RQ, Sheng J, Liu C, Ma H, Wang HX, Li JH, Gao XH, Wan YS, Chen HD. MiR-155, a potential serum marker of extramammary Paget’s disease. BMC Cancer. 2018; 18:1078. DOI: 10.1186/s12885-018-4994-1. PMID: 30458743. PMCID: PMC6247506.

28. Jia L, Liu W, Cao B, Li H, Yin C. MiR-507 inhibits the migration and invasion of human breastcancer cells through Flt-1 suppression. Oncotarget. 2016; 7:36743–36754. DOI: 10.18632/oncotarget.9163. PMID: 27167339. PMCID: PMC5095036.

29. Wang LH, Lin CY, Liu SC, Liu GT, Chen YL, Chen JJ, Chan CH, Lin TY, Chen CK, Xu GH, Chen SS, Tang CH, Wang SW. CCL5 promotes VEGF-C production and induces lymphangiogenesis by suppressing miR-507 in human chondrosarcoma cells. Oncotarget. 2016; 7:36896–36908. PMID: 27166194. PMCID: PMC5095047.

30. Gross C, Dubois-Pot H, Wasylyk B. The ternary complex factor Net/Elk-3 participates in the transcriptional response to hypoxia and regulates HIF-1 alpha. Oncogene. 2008; 27:1333–1341. DOI: 10.1038/sj.onc.1210736. PMID: 17704799.

31. Wasylyk C, Zheng H, Castell C, Debussche L, Multon MC, Wasylyk B. Inhibition of the Ras-Net (Elk-3) pathway by a novel pyrazole that affects microtubules. Cancer Res. 2008; 68:1275–1283. DOI: 10.1158/0008-5472.CAN-07-2674. PMID: 18316589.

32. Lee JH, Hur W, Hong SW, Kim JH, Kim SM, Lee EB, Yoon SK. ELK3 promotes the migration and invasion of liver cancer stem cells by targeting HIF-1α. Oncol Rep. 2017; 37:813–822. DOI: 10.3892/or.2016.5293. PMID: 27959451.

33. Park JH, Kim KP, Ko JJ, Park KS. PI3K/Akt/mTOR activation by suppression of ELK3 mediates chemosensitivity of MDA-MB-231 cells to doxorubicin by inhibiting autophagy. Biochem Biophys Res Commun. 2016; 477:277–282. DOI: 10.1016/j.bbrc.2016.06.057. PMID: 27301639.

Fig. 1
LINC00525 was highly expressed in CRC and increased LINC00525 expression predicted poor prognosis in CRC patients. (a) The expression level of LINC00525 was analyzed in CRC tissues compared with normal tissues from GSE21510. (b) qRT-PCR analysis of LINC00525 expression in CRC tissues (n=60) and normal human intestinal tissues (n=60) (shown as log10 (2−ΔΔCT)). (c) The LINC00525 expression level was quantified by qRT-PCR analysis in CRC tissues of oxaliplatin sensitive patients (OxS) and oxaliplatin resistant patients (OxR). (d) qRT-PCR analysis of the relative expressions of LINC00525 in five CRC cell lines (Caco2, SW480, SW620, HCT116, and HT29) and a normal human intestinal epithelial cell line (HIEC). (e) qRT-PCR analysis of the relative expressions of LINC00525 in sphere-forming HCT116 and HT29 compared to parental HCT116 and HT29 cells. (f) Kaplan–Meier’s analysis of the correlation between LINC00525 expression and the overall survival of CRC patients. *p<0.05, **p<0.01, ***p<0.001.
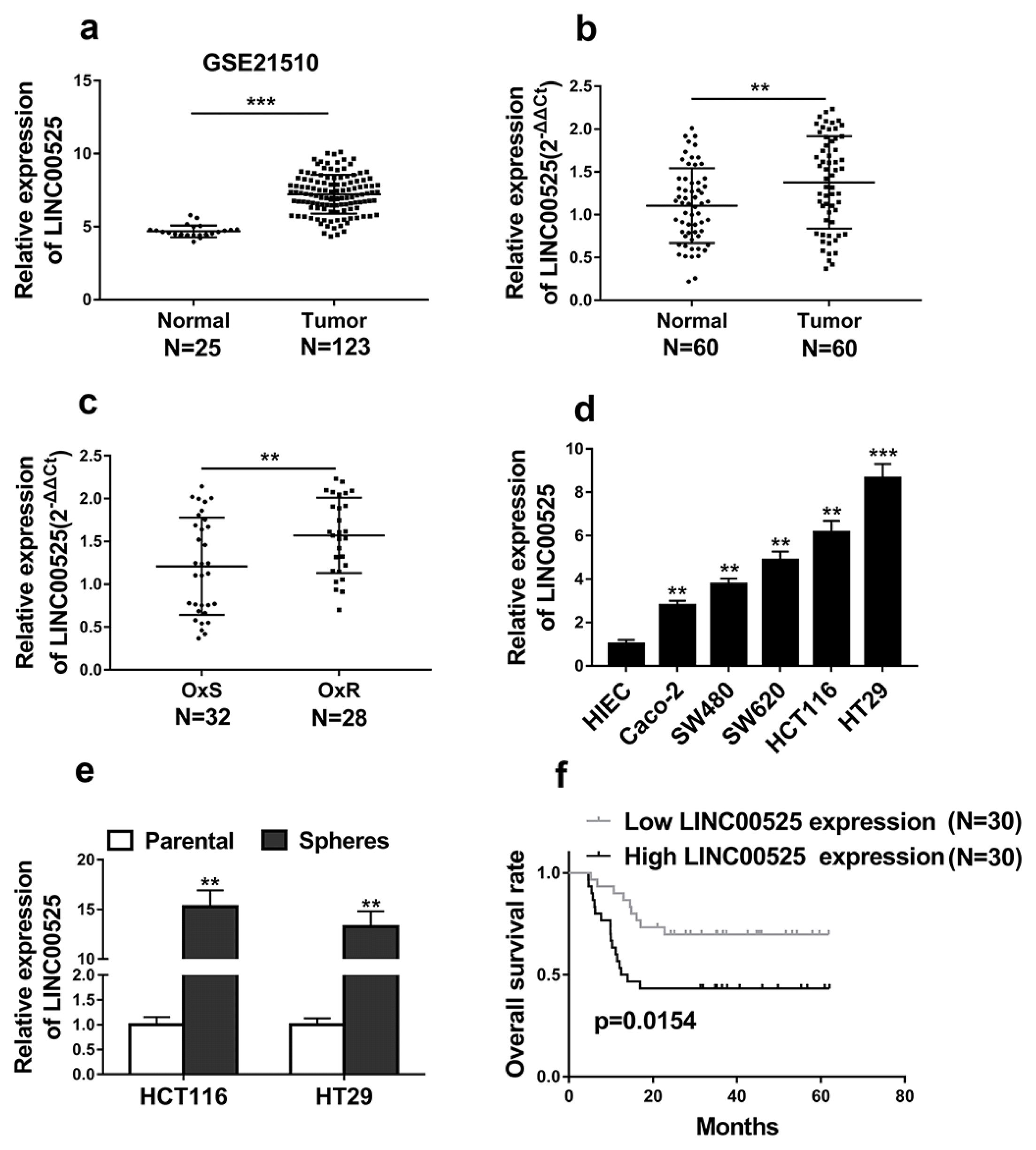
Fig. 2
Silencing of LINC00525 inhibited the stemness and oxaliplatin resistance of colorectal cancer. (a) The relative expression levels of LINC00525 in HCT116 and HT29 cells transfected with LINC00525 shRNAs (sh-LINC00525-1, sh-LINC00525-2) or empty vector (sh-CTR) were detected by qRT-PCR. (b) Knockdown of LINC00525 in HCT116 and HT29 cells significantly decreased the capacity to form tumorspheres. (c~e) The levels of CD44, SOX2 and OCT4 were determined by qRT-PCR and western blot in HCT116 and HT29 cells transfected with LINC00525 shRNAs (sh-LINC00525-1, sh-LINC00525-2) or empty vector (sh-CTR). (f) The relative expression levels of LINC00525 in oxaliplatin resistant HCT116 (HCT116-OxR) and HT29 (HT29-OxR) cells transfected with LINC00525 shRNAs (sh-LINC00525-1, sh-LINC00525-2) or empty vector (sh-CTR) were detected by qRT-PCR. (g~i) Knockdown of LINC00525 significantly inhibited CRC-OxR cells growth under oxaliplatin treatment. Cell viabilities were determined by MTT and colony formation assays. (j) Inhibition of LINC00525 significantly promoted CRC-OxR cell apoptosis. Cell apoptosis was determined by DNA fragmentation assay. (k) The apoptotic markers p53, Caspase-3, Bcl-2 in oxaliplatin resistant HCT116 (HCT116-OxR) and HT29 (HT29-OxR) cells transfected with LINC00525 shRNAs (sh-LINC00525-1, sh-LINC00525-2) or empty vector (sh-CTR) were detected by western blotting. The data represent as mean±SD from three independent experiments. *p<0.05, **p<0.01.
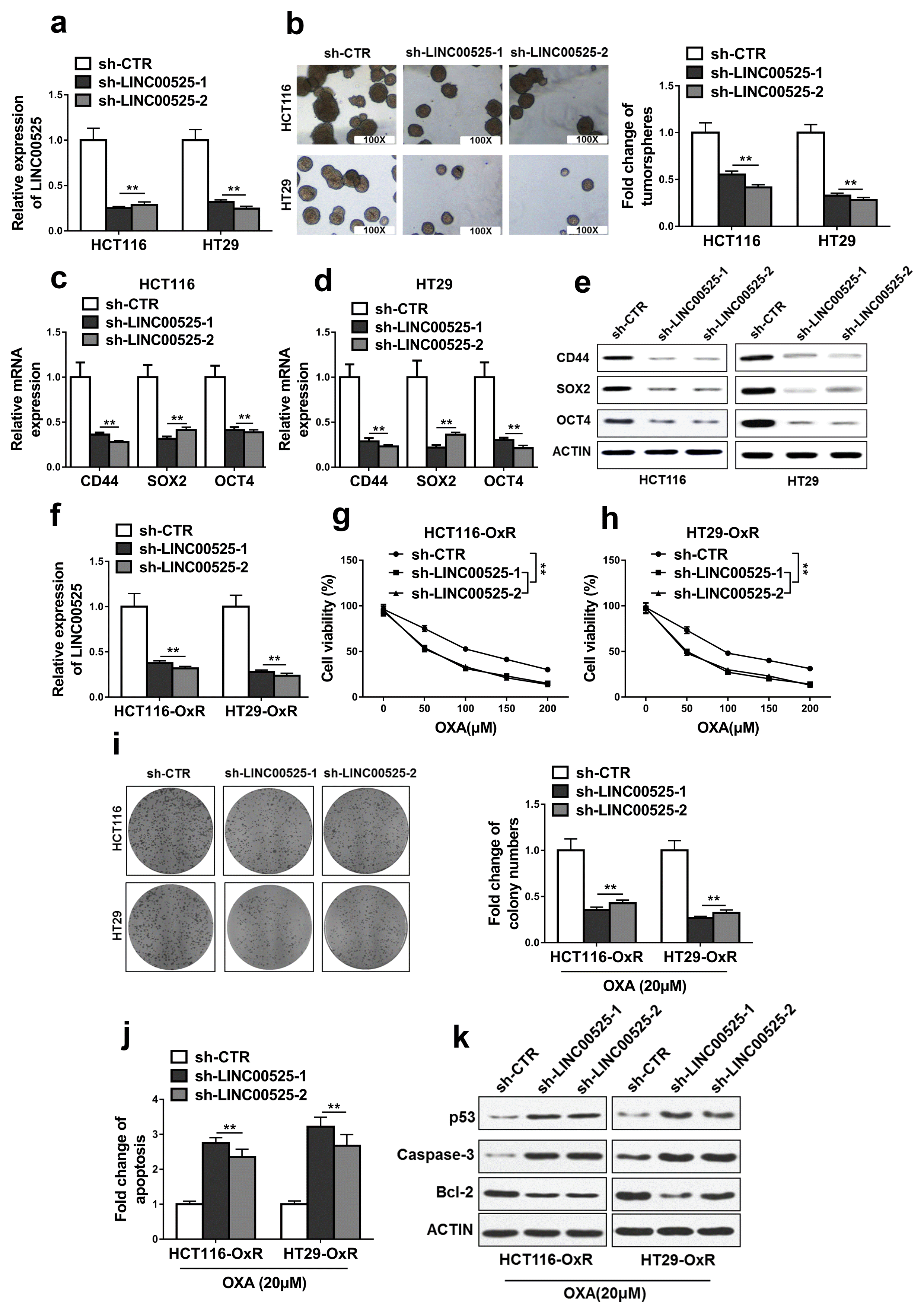
Fig. 3
MiR-507 was a direct target of LINC00525. (a) The prediction for miR-507 binding sites on LINC00525 transcript and schematic of luciferase reporter vector constructs LINC00525 wild-type (LINC00525 wt) and the miR-507-binding-site mutated (LINC00525 mt) one. (b, c) The luciferase activities in HCT116 and HT29 cells co-transfected with miR-507 or miR-NC mimics and luciferase reporters containing LINC00525 wt or LINC00525 mt. Data are presented as the relative ratio of hRluc luciferase activity to hluc+ luciferase activity. (d) The relative expression levels of miR-507 in HCT116 and HT29 cells transfected with LINC00525 shRNAs (sh-LINC00525-1, sh-LINC00525-2) or empty vector (sh-CTR) were detected by qRT-PCR. (e) The relative expression levels of miR-507 in HCT116 and HT29 cells transfected with LINC00525 plasmid (pSin-LINC00525) or empty vector (pSin-vec) were detected by qRT-PCR. (f, g) Lysates from HCT116 and HT29 cells were incubated with in vitro-synthesized biotin-labeled sense or antisense DNA probes against LINC00525 for biotin pull-down assay, followed by qRT-PCR analysis. (h) The relative expression levels of LINC00525 in HCT116 and HT29 cells transfected with miR-507 mimics (miR-507) or mimics negative control (miR-NC) were detected by qRT-PCR. The data represent as mean±SD from three independent experiments. *p<0.05, **p<0.01, ns indicates no significance.
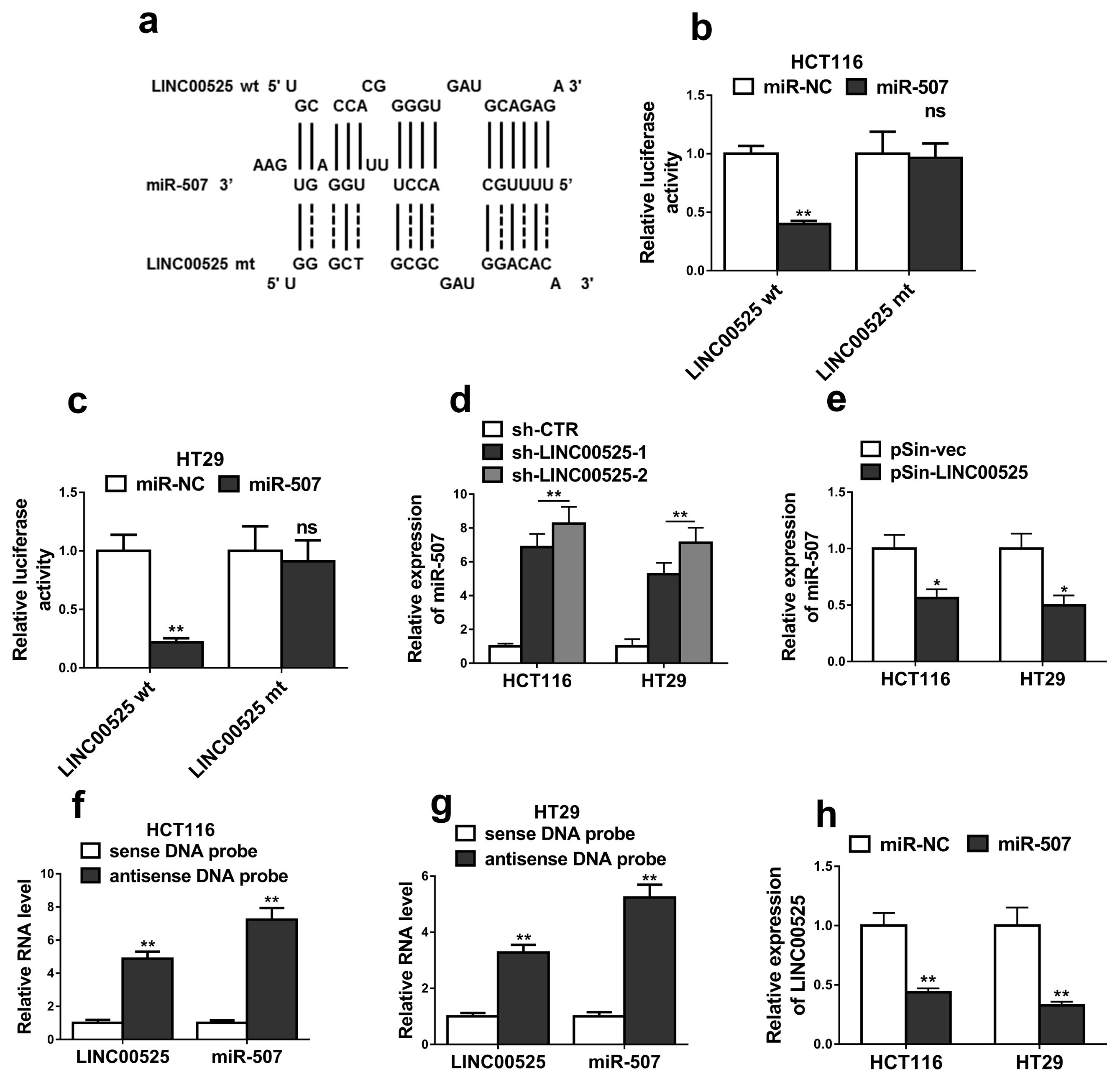
Fig. 4
MiR-507 suppressed the stemness and oxaliplatin resistance of colorectal cancer. (a) The relative expression levels of miR-507 in HCT116 and HT29 cells transfected with miR-507 mimics (miR-507) or mimics negative control (miR-NC) were detected by qRT-PCR. (b) Overexpression of miR-507 in HCT116 and HT29 cells significantly decreased the capacity to form tumorspheres. (c~e) The levels of CD44, SOX2 and OCT4 were determined by qRT-PCR and western blotting in HCT116 and HT29 cells transfected with miR-507 mimics (miR-507) or mimics negative control (miR-NC). (f) The relative expression levels of miR-507 in oxaliplatin resistant HCT116 (HCT116-OxR) and HT29 (HT29-OxR) cells transfected with miR-507 mimics (miR-507) or mimics negative control (miR-NC) were detected by qRT-PCR. (g~i) Overexpression of miR-507 significantly inhibited CRC-OxR cells growth. Cell viabilities were determined by MTT and colony formation assays. (j) Overexpression of miR-507 increased CRC-OxR cells apoptosis. Cell apoptosis was determined by DNA fragmentation assay. (k) The apoptotic markers p53, Caspase-3, Bcl-2 in oxaliplatin resistant HCT116 (HCT116-OxR) and HT29 (HT29-OxR) cells transfected with miR-507 mimics (miR-507) or mimics negative control (miR-NC) were detected by western blotting. The data represent as mean±SD from three independent experiments. *p<0.05, **p<0.01, ***p<0.001.
Fig. 5
ELK3 was a direct target of miR-507. (a) The sequences of binding sites between miR-507 and ELK3 3′UTR, including wild-type (ELK3 3′UTR (WT)) and mutant (ELK3 3′UTR (MT)). (b, c) Luciferase reporter assay was performed in the cells co-transfected with miR-507 or miR-NC mimics and plasmids containing ELK3 3′UTR (WT) or ELK3 3′UTR (MT). (d) ELK3 mRNAs in biotinylated miRNA/target mRNA complex was detected using qRT-PCR. Biotinylated miR-507 (bio-miR-507) or biotinylated control random RNA (bio-miR-NC) was used to pulldown ELK3 mRNA in the complex. (e~g) The relative expression levels of ELK3 in HCT116 and HT29 cells transfected with indicated microRNA mimics or microRNA inhibitors were detected by qRT-PCR and western blotting. (h~j) The relative expression levels of ELK3 were detected by qRT-PCR and western blot. HCT116 and HT29 cells were transfected with LINC00525 shRNAs (sh-LINC00525-1, sh-LINC00525-2) or empty vector (sh-CTR) and LINC00525 plasmid (pSin-LINC00525) or empty vector (pSin-vec). Data are representative of three or more independent experiments. The data represent as mean±SD from three independent experiments. *p<0.05, **p<0.01, ns indicates no significance.
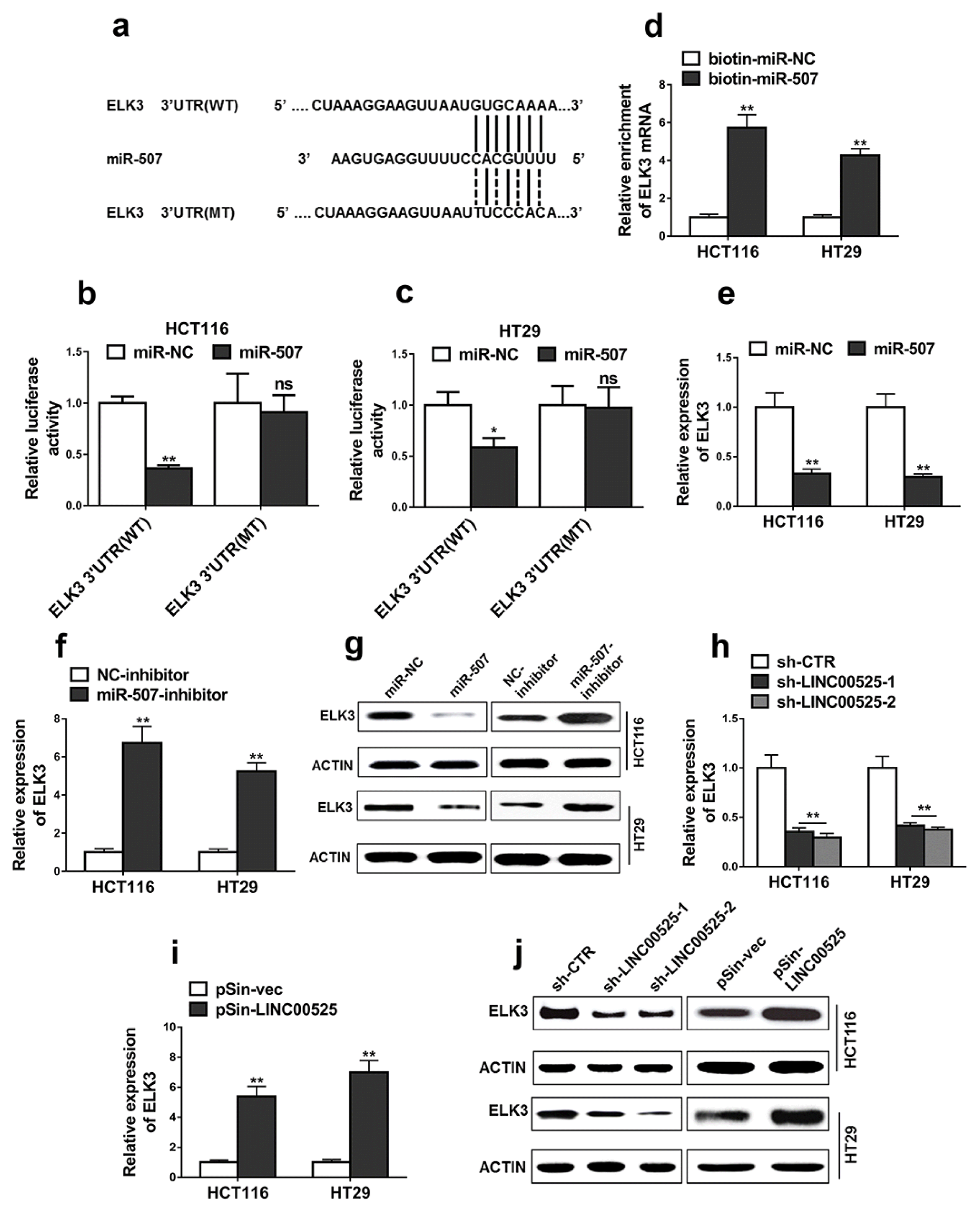
Fig. 6
Restoration of ELK3 expression abrogated the effects of LINC00525 knockdown. (a, b) Expression levels of ELK3 were determined by qRT-PCR and western blot in the parental HCT116 cells. (c) The capacity to form tumorspheres of these parental HCT116 cells was determined by tumorsphere formation assay. (d, e) mRNA and protein levels of CD44, SOX2 and OCT4 in these parental HCT116 cells were determined by qRT-PCR and western blotting. (f~j) Oxaliplatin resistant HCT116 (HCT116-OxR) cells subjected to co-transfection with empty plasmids (sh-CTR+pSin-vec), LINC00525 shRNA and empty plasmid (sh-LINC00525-1+pSin-vec) or LINC00525 shRNA and ELK3 overexpression plasmid (sh-LINC00525-1+pSin-ELK3). (f, g) Expression levels of ELK3 were determined by qRT-PCR and western blotting in the oxaliplatin resistant HCT116 (HCT116-OxR) cells. (h, i) MTT and colony formation assays were used to determine cell growth of the oxaliplatin resistant HCT116 (HCT116-OxR) cells. (j) Apoptosis levels of these oxaliplatin resistant HCT116 (HCT116-OxR) cells were measured by DNA fragmentation assay. (k) The apoptotic markers p53, Caspase-3, Bcl-2 in these oxaliplatin resistant HCT116 (HCT116-OxR) cells were detected by western blotting. The Data represent as mean±SD from three or more independent experiments. *p<0.05, **p<0.01, ***p<0.001.




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download