Introduction
Materials and Methods
Isolation and culture conditions of human MSC and Ec
Isolation and characterization of HSC from human umbilical cord blood (UCB)
Generation of the 3D cultivation system by magnetic levitation
Evaluation of OMS: volume, sphericity, aggregation and viability
Histochemical characteristics of spheres
Functional evaluation of HSC
Statistical analysis
Results
Isolation, culture and characterization of cell populations: MSC, Ec and HSC
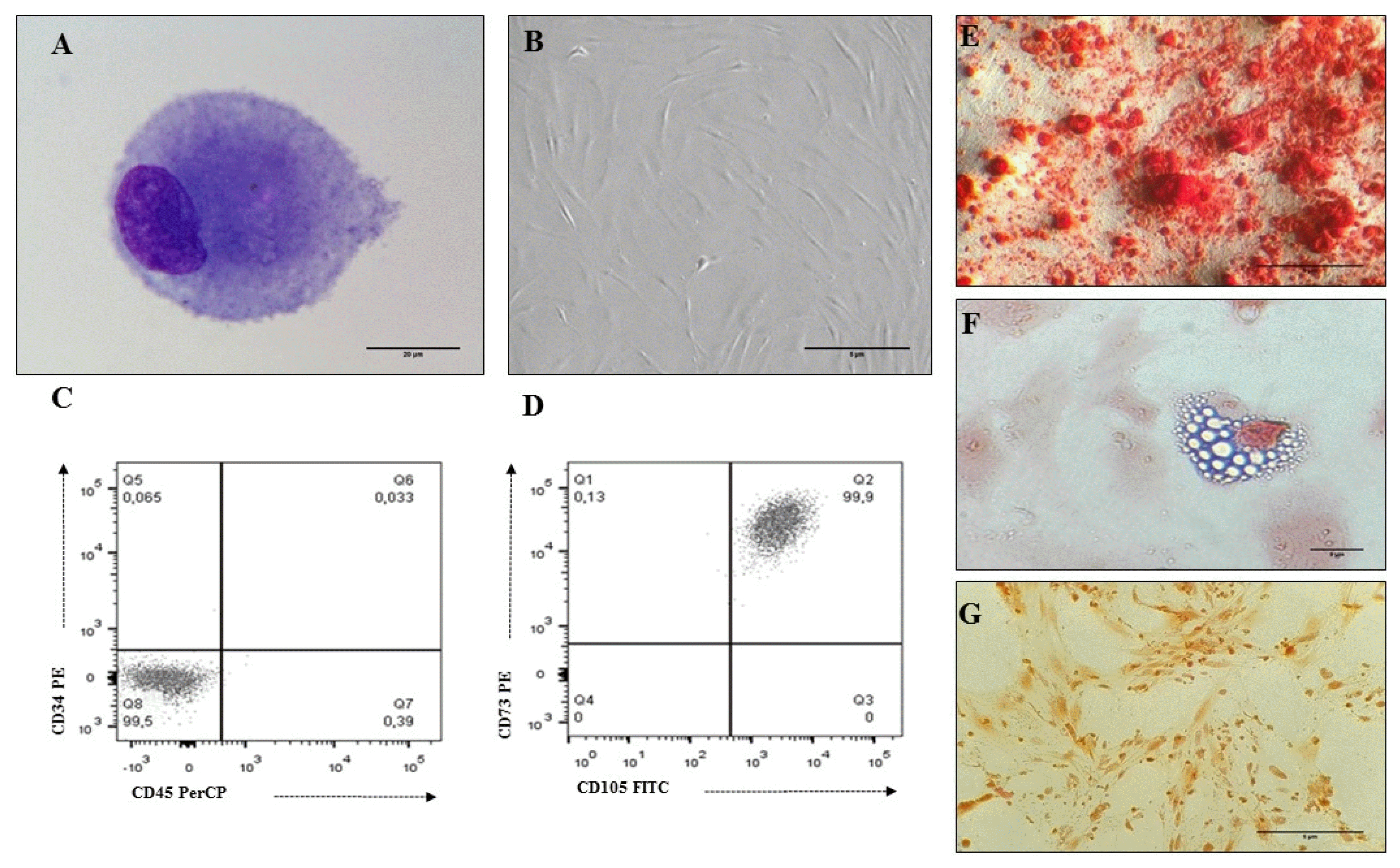 | Fig. 1MSC characteristics isolated from human BM. (A) MSC Morphology (3 passage): cells with an eccentric nucleus, loose chromatin and nucleoli; presence of granular basophilic cytoplasm (×100 Cythospin, hematoxylin and eosin stain, optical microscopy. Scale bar: 20 μm), (B) MSC Morphology (3 passage): Fibroblastoid cells associated with MSC (×20, inverted microscopy. Scale bar: 5 μm), (C, D) Immunophenotype (3 passage): MSCs do not express hematopoietic antigens (CD34 and CD45) and co-express CD73 and CD105, (E) Osteogenic differentiation (×20, red alizarin stain, inverted microscopy. Scale bar: 5 μm), (F) Adipogenic differentiation (×40, sudan black stain, inverted microscopy. Scale bar: 5 μm), (G) Chondrogenic differentiation (×20, safranin O stain, inverted microscopy. Scale bar: 5 μm). |
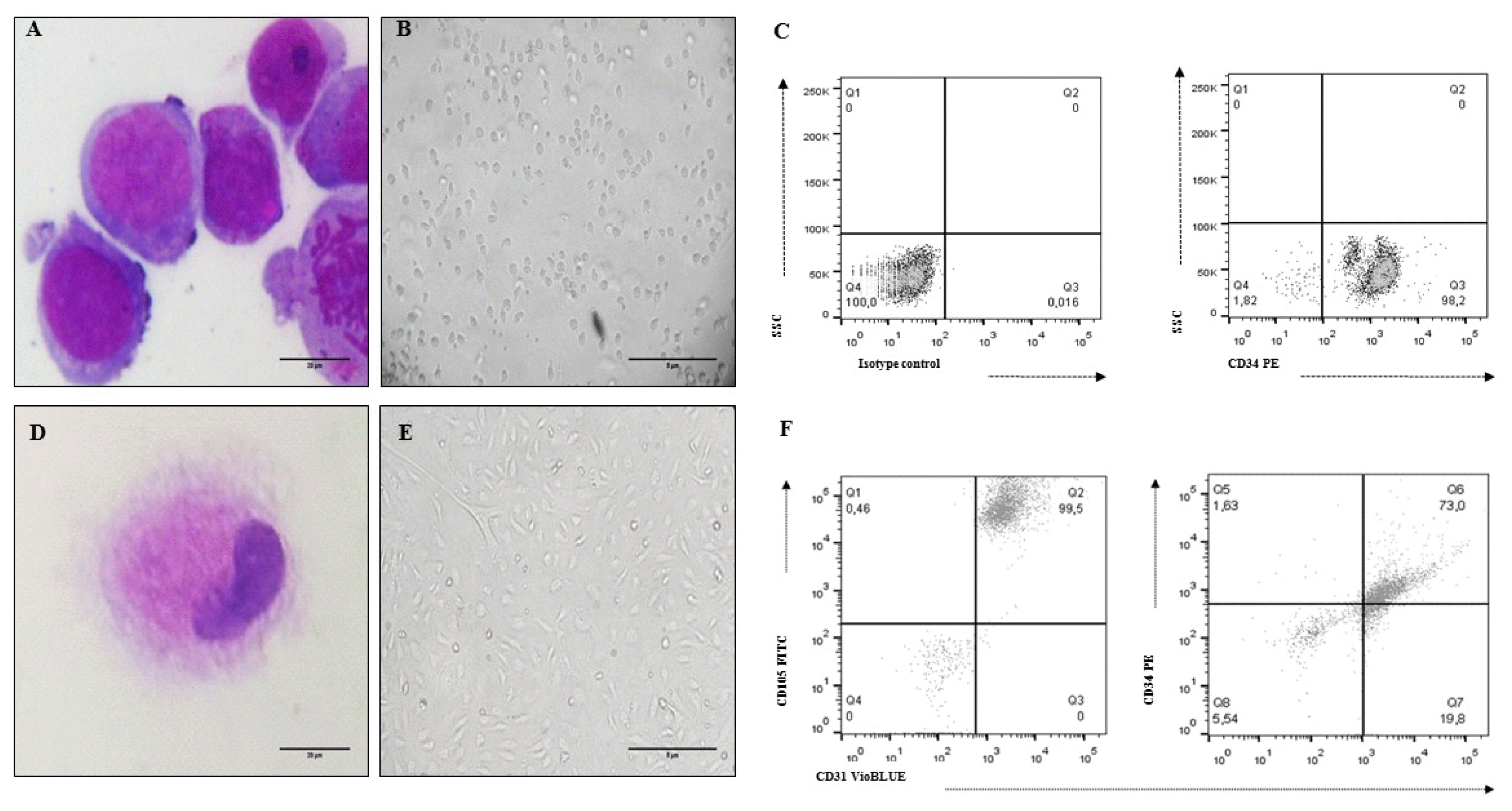 | Fig. 2UCB-HSC and Ec characteristics (cell line CC-2811. Lonza®). (A) HSC Morphology: cells with a predominant nucleus, nucleoli and basophil cytoplasm (×100 Cythospin, hematoxylin and eosin stain, optical microscopy. Scale bar: 20 μm), (B) Non-adherent cells compatible with UCB-HSC (×20, inverted microscopy. Scale bar: 5 μm), (C) HSC phenotype: CD34+, (D) Ec Morphology: cells with metacentric nucleus and abundant cytoplasm (×100 Cythospin, hematoxylin and eosin stain, optical microscopy. Scale bar: 20 μm), (E) Adherent cells compatible with Ec (×20, inverted microscopy. Scale bar: 5 μm), (F) Ec phenotype: CD34+, CD31+, CD105+. |
Table 1
Table 2
Table 3
Relationship between cell populations and nanoparticles to obtain OMS
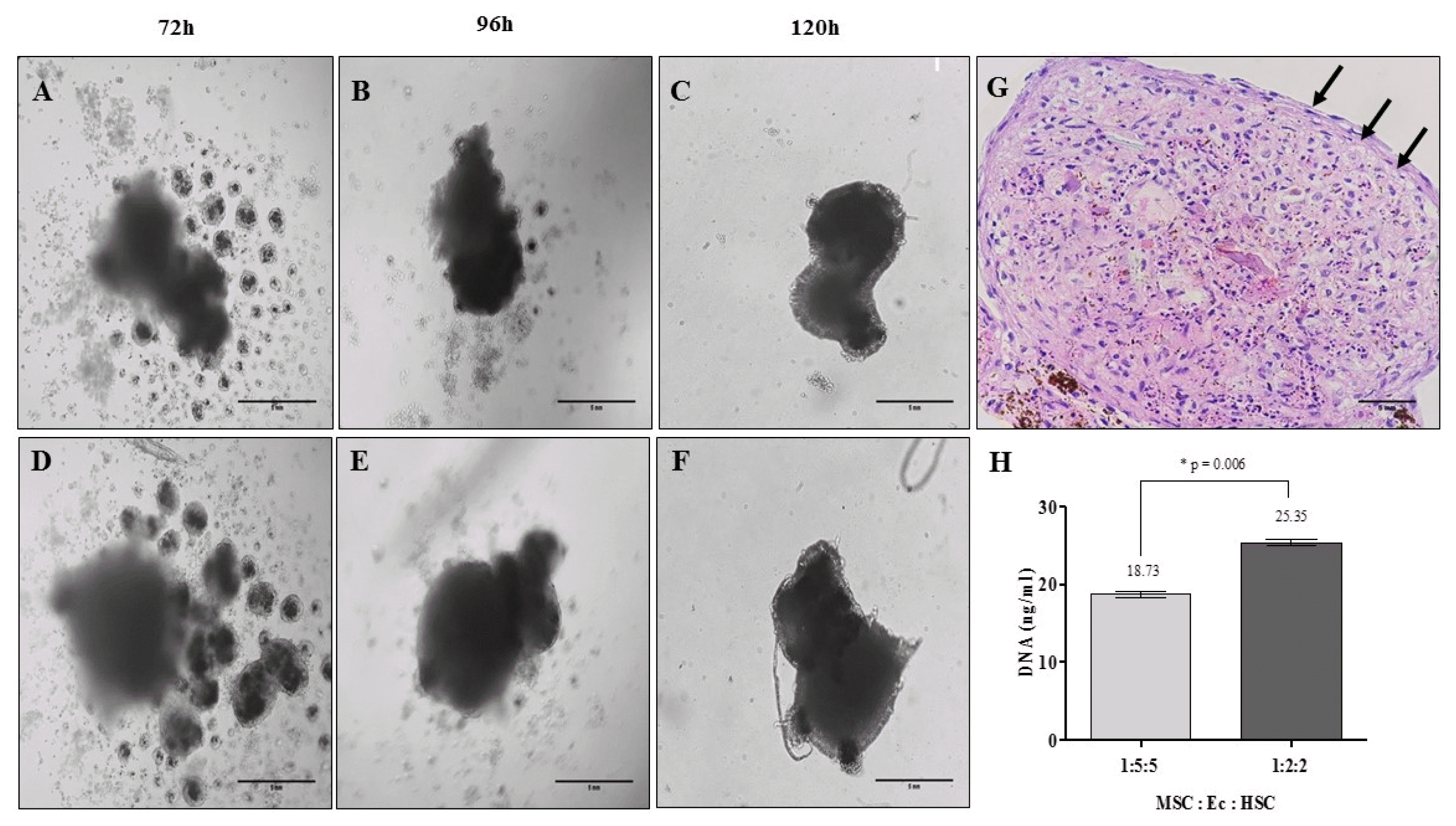 | Fig. 3Organotypic multicellular spheroid (OMS) formation after 5 days: time vs. cell density. (A~C) Relation 1:5:5 MSC:Ec:HSC (×20, inverted microscopy. Scale bar: 5 mm), (D~F) Relation 1:2:2 MSC:Ec:HSC (Total cell density 1×105 cells) (×20, inverted microscopy. Scale bar: 5 mm). (G) Morphology of OMS after 5 days of culture. It shows cells that are organized in the periphery of the structure demilitarizing the sphere (Relation 1:2:2 MSC:Ec:HSC. ×40, hematoxylin & eosin. Scale bar: 5 mm). (H) DNA quantification in the spheres of the two experimental conditions (p<0.05). |
General characteristics of multicellular spheres
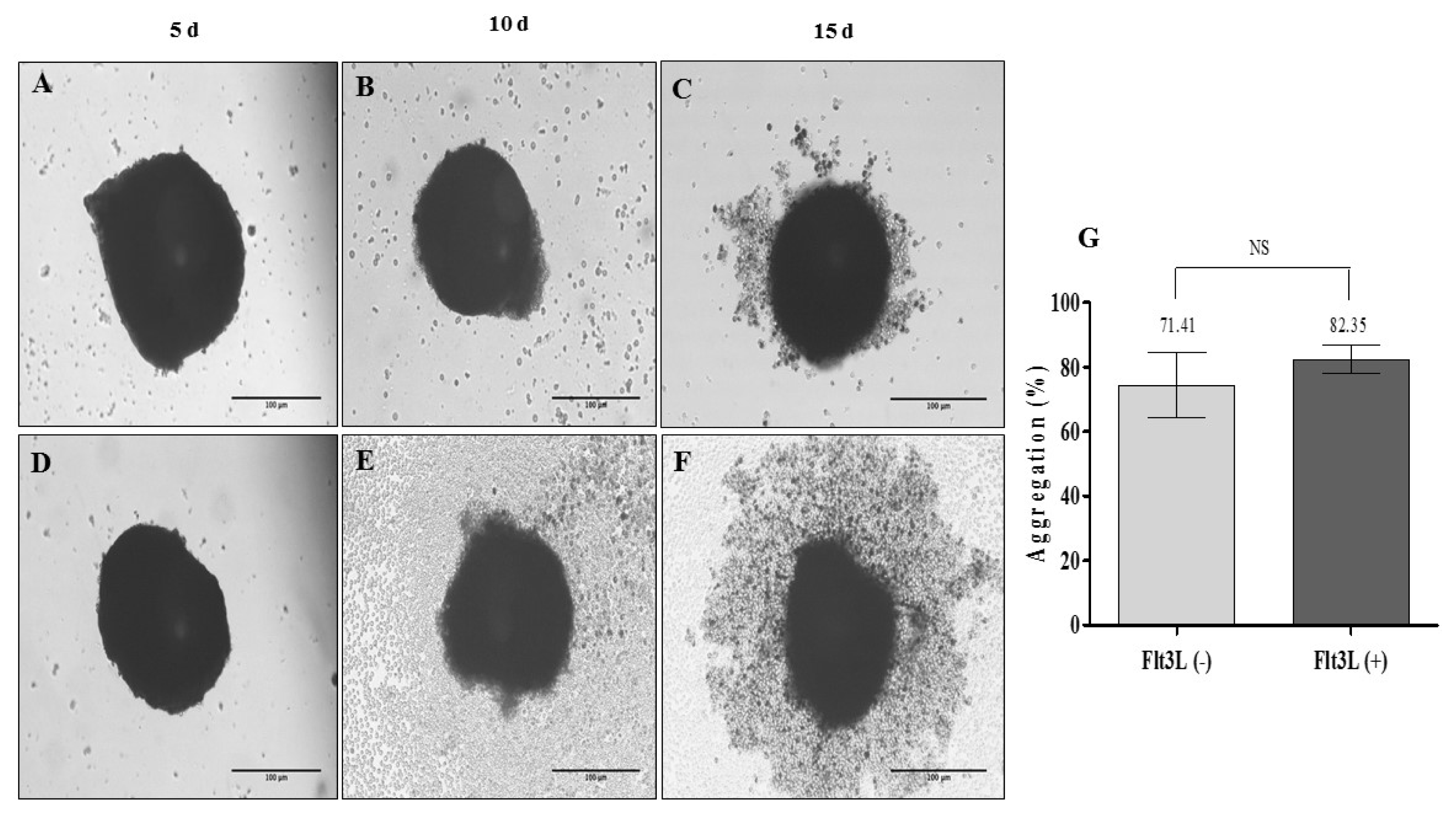 | Fig. 4Organotypic multicellular spheroid (OMS) (MSC: Ec: HSC of 1:2:2). (A~C) 3D culture without addition of exogenous cytokines (Flt3L −) (×20, inverted microscopy. Scale bar: 100 μm), (D~F) 3D culture with 100 ng/ml Flt3-L (Flt3L +) (×20, inverted microscopy. Scale bar: 100 μm). (G) Percentage of aggregation of OMS cultivated without and with Flt3L (p>0.05). |
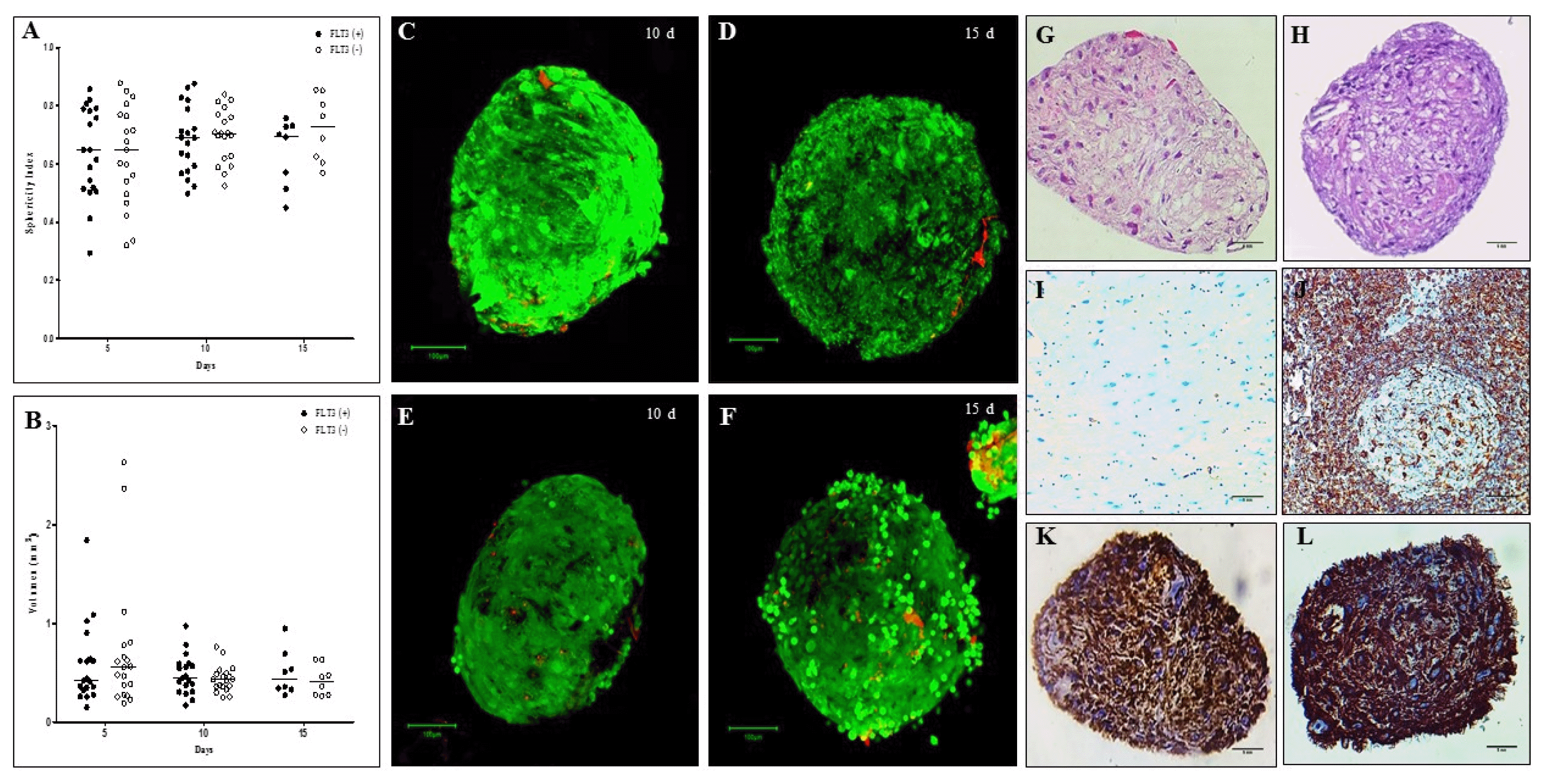 | Fig. 5Sphericity, volume, viability and histological evaluation of organotypic multicellular spheroid (OMS). (A) Sphericity Index, non significant differences were observed between the spheres cultivated without and with Flt3L, (B) Volumen, similarly there are no differences between the experimental conditions (p>0.05). 3D structures were treated with exogenous Flt3L (100 ng/ml) (C, D) or without cytokine (E–F). Viability was evaluated with LIVE/DEADTM Cell Imaging Kit (Invitrogen) and confocal microscope FV1000 (Olympus) using an UPLSAPO 20× (1.35 NA oil immersion objective), 488 nm argon laser line for green fluorescence (living cells) and HeNe 543 nm laser for red fluorescence (dead cells). Data analysis was performed with the FlowView and Fiji software (green cells: alive, red cells: dead) (Scale bar: 100 um). For histological evaluation, after 15 days of culture, cells without pyknotic nuclei are observed in the OMS (G) without Flt3L (×40, hematoxylin & eosin. Scale bar: 5 mm) and (H) with Flt3L (×40, hematoxylin & eosin. Scale bar: 5 mm) (I) Negative control of vimentin (brain tissue) (×40. Scale bar: 5 mm), (J) Positive control of vimentin (lymph node) (×40. Scale bar: 5 mm), (K) Vimentina in OMS without Flt3L (×40. Scale bar: 5 mm) and (L) OMS with Flt3L (×40. Scale bar: 5 mm). |
Histological evaluation of the spheres
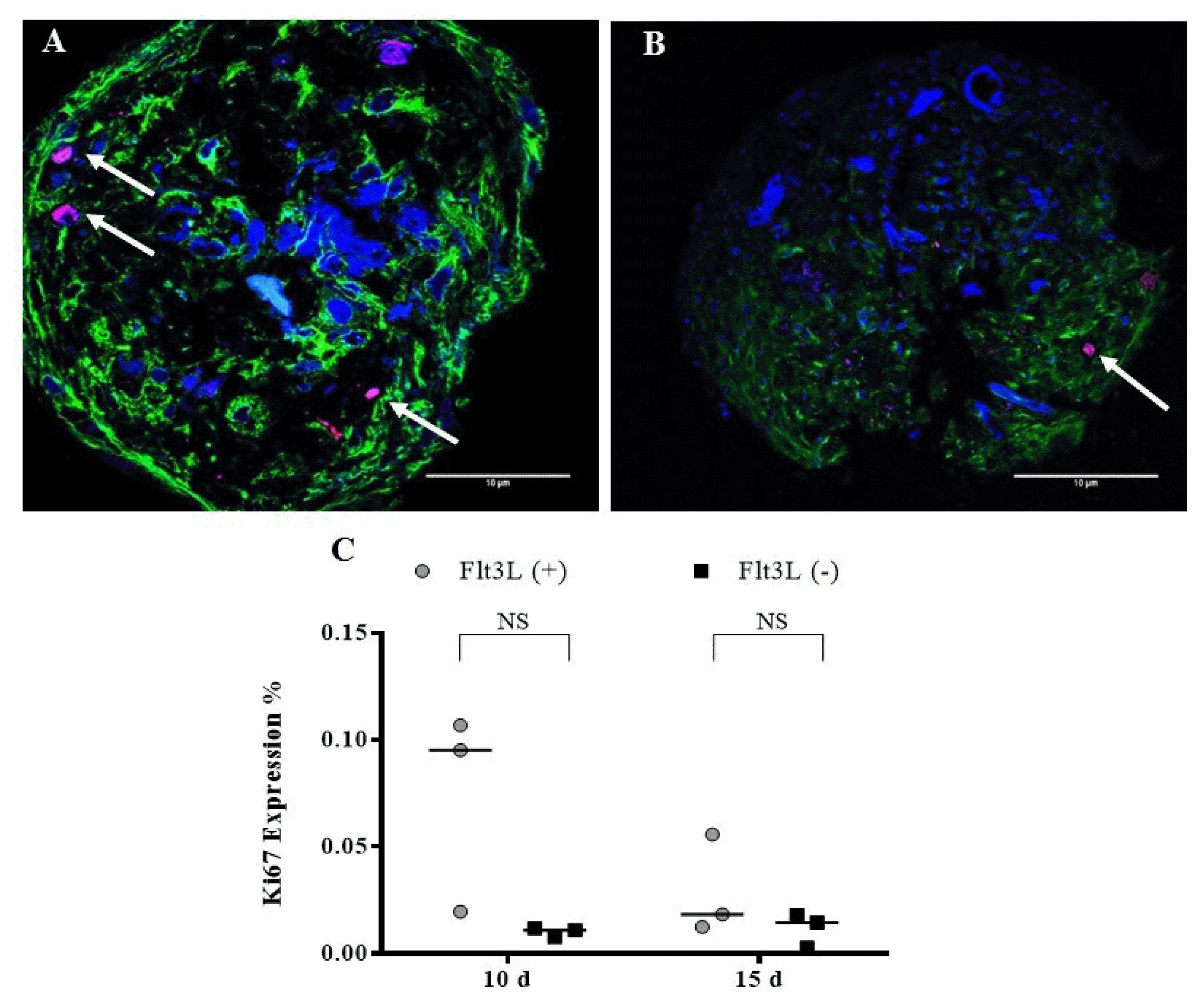 | Fig. 6Ki67 expression. OMS were cultivated with (A) Flt3L (+) and (B) without the cytokine Flt3L (cells expressing Ki67 antigen are indicated with the arrows). Cuts of the spheres were imaged with laser scanning confocal microscope FV1000 (Olympus) using an UPLSAPO 20×, 1.35 NA oil immersion objective. A 405 nm diode laser was used for DAPI (nuclei), 488 nm argon laser line for Alexa-Fluor 488 nm (α-tubulin) and Argon 515 nm laser for alexa-fluor 514 nm (Ki67). Data analysis was performed with the FlowView and ImageJ software (green: α-tubulin, blue: DAPI, magenta: Ki67) (Scale bar: 10 μm). (C) Percentage of Ki67 expression after 10 and 15 d. |
Functional evaluation of HSC after cultivation in multicellular spheres
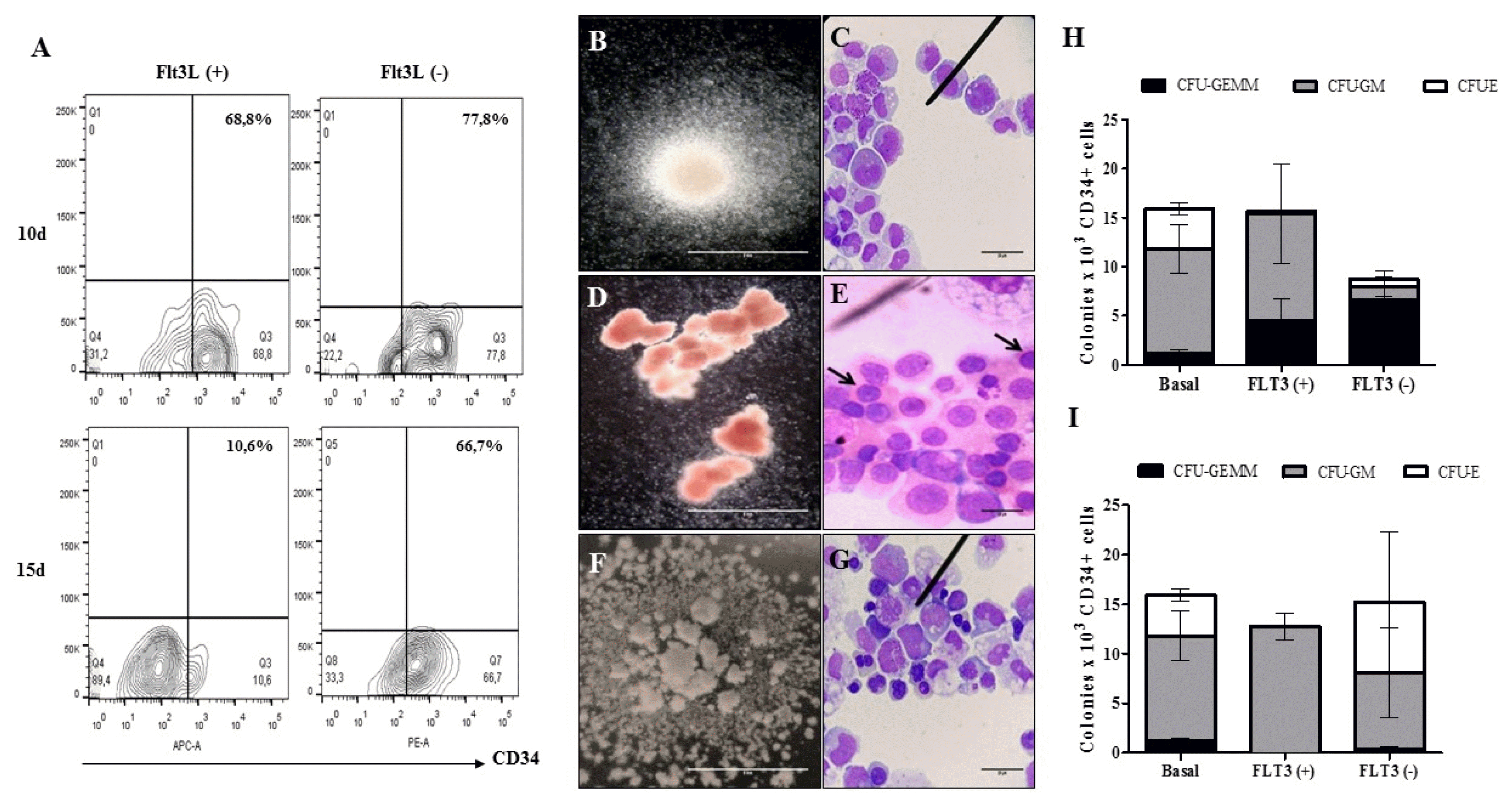 | Fig. 7Multipotent capacity of HSC after cultivation in OMS. (A) Dot plot representative of the recovery percentages of CD34+ cells after the disintegration of the multicellular spheres. The ability of HSC to generate (B) CFU-GM (colony forming unit - granulocyte/monocyte progenitor. ×10, inverted microscope. Scale bar: 5 mm) (C) Immature granulocytic and monocytic cells: promyelocytes, myelocytes, promonocytes and bands cells (×100, hematoxylin & eosin. Scale bar: 20 μm), (D) CFU-E (colony forming unit - erythroid progenitor. ×10, inverted microscope. Scale bar: 5 mm), (E) Immature erythroid cells: orthochromatic normoblasts are indicated (×100, hematoxylin & eosin. Scale bar: 20 μm), (F) CFU-GEMM (colony forming unit – granulocyte, erythrocyte, monocyte, megakaryocyte. ×10, inverted microscope. Scale bar: 5 mm) and (G) Immature cells of myeloid lineage: myeloblasts, promyelocytes, normoblasts, promonocytes (×100, hematoxylin & eosin. Scale bar: 20 μm) was evaluated after 10 d (H) and 15d (I) (n=1) (p>0.05). |




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download