Abstract
Background and Objectives
Embryonic stem (ES) cells have pluripotent ability to differentiate into multiple tissue lineages. SIRT1 is a class III histone deacetylase which modulates chromatin remodeling, gene silencing, cell survival, metabolism, and development. In this study, we examined the effects of SIRT1 inhibitors on the hematopoietic differentiation of mouse ES cells.
Methods and Results
Treatment with the SIRT1 inhibitors, nicotinamide and splitomicin, during the hematopoietic differentiation of ES cells enhanced the production of hematopoietic progenitors and slightly up-regulated erythroid and myeloid specific gene expression. Furthermore, treatment with splitomicin increased the percentage of erythroid and myeloid lineage cells.
Conclusions
Application of the SIRT1 inhibitor splitomicin during ES cell differentiation to hematopoietic cells enhanced the yield of specific hematopoietic lineage cells from ES cells. This result suggests that SIRT1 is involved in the regulation of hematopoietic differentiation of specific lineages and that the modulation of the SIRT1 activity can be a strategy to enhance the efficiency of hematopoietic differentiation.
ES cells can give rise to all cell types derived from the three embryonic germ layers and are able to differentiate into a variety of specialized cells (1). During the last few decades, numerous researchers have given extensive efforts to elucidate the underlying molecular mechanisms that control the self-renewal and differentiation of ES cells (2). Furthermore, significant attention has been focused on the use of stem cells to obtain specialized cells for treating a variety of diseases.
Generating transplantable hematopoietic cells from ES cells in vitro will be one of the main ultimate goals of ES cell-based cell replacement therapy (3). Under the appropriate conditions in culture such as in the absence of a feeder layer and LIF, ES cells can be differentiated into embryonic bodies (EBs). EBs contain several different cell types including endothelial, muscle, neuronal and hematopoietic progenitors (4). In vitro hematopoietic differentiation of mouse embryonic stem (mES) cells have been examined in co-culture with stromal cells, in chemically-defined suspension media in the presence of hematopoiesis factors, or in methylcellulose-based semisolid media containing cytokines (5). In the co-culture system with stromal cells such as the ST2 and OP9 cell lines, myeloid and lymphoid precursors were simultaneously obtained from ES cells. However, this method has a limitation because of the possible contamination of the feeder cells (6). Hematopoietic differentiation of EBs can be effectively induced by stimulation with the appropriate cytokines. In the early studies on hematopoietic differentiation, only red blood cells were detected in EBs. In 1991, it was reported that EBs cultured in the presence of IL-3 in semisolid media differentiated into macrophages, neutrophils, and mast cells (7). Differentiation in the presence of growth factors specific for mesoderm (BMP4, FGF and activin A) and blood formation (VEGF, SCF, IL-3, IL-6, G-SCF and TPO) promotes hematopoiesis within EBs (8).
Gene expression analysis of differentiating ES cells demonstrated that several genes are implicated during hematopoietic differentiation. Brachyury, a mesodermal marker gene, is indispensable for mesodermal formation (9). Subsequently, Flk1 is necessary for blood island formation and is expressed in hemangioblasts which are common embryonic endothelial and hematopoietic precursors (10). In the transition from mesoderm to hematopoietic lineage commitment, transcription factor Scl is indispensable for the development of all hematopoietic lineages (11). The GATA gene family of transcription factors, especially GATA1 and GATA2, have key roles in the positive regulation of erythroid and megakaryocyte development (12). βH1 and β-major are globin genes that are expressed at the developmental stage of the erythroid (4, 5). As a primitive and definitive marker of erythroid development, analysis of the globin gene expression enables a useful assessment of differentiation into either the primitive or definitive lineages (7). PU.1, a myeloid specific marker, is an extensively studied hematopoietic transcription factor that is essential for embryonic myeloid development (13). Although hematopoietic differentiation of ES cells has been studied for several decades, understanding the mechanisms regulating lineage fate determination is still limited. Therefore, deciphering the mechanisms involved in hematopoietic differentiation and controlling the process has immense therapeutic promise considering the impact of blood diseases in human health.
The class III histone deacetylases (HDAC), also known as the sirtuins, are a member of the sirtuin family of proteins (SIRT1~7) and homologs of Sir2 (silent information regulator 2) in yeast. SIRT1 shares a domain of about 275 amino acids with other sirtuins and uses NAD+ to mediate the deacetylation of histone and non-histone proteins and consequently contributes to regulating various cellular functions including control of gene expression, differentiation, metabolism, aging, and tumor suppression (14). SIRT1 is expressed at high levels in mouse embryonic day 4.5 embryos. Although its expression is down-regulated during the subsequent embryogenesis, a high level of expression remains detectable at embryonic day 18.5 (15). It was suggested that SIRT1 is responsible for the greater functionality of endothelia cells derived from ES cells and iPS cells compared to adult endothelial cells (16). In a comparative study using SIRT1+/+, SIRT1+/−, and SIRT1−/− ES cells, SIRT1 was shown to contribute to embryonic and adult hematopoiesis (17).
Recently, small molecules have emerged as essential tools for understanding and regulating stem cells and manipulating stem cell fate. Nicotinamide, a form of vitamin B3 serving as a precursor of nicotinamide adenine dinucleotide (NAD), is a well-established potent inhibitor of SIRT1 (18, 19). Giammona et al. showed that maturation of megakaryocytes in vitro can be dramatically enhanced by adding nicotinamide (20). However, another study reported that nicotinamide delayed differentiation and increased the engraftment efficacy of cord blood–derived human CD34+ cells cultured with cytokines (21). Splitomicin is derived from β-naphthol and is an inhibitor of SIRT1 and SIRT2 (22, 23). Previous studies have shown that splitomicin inhibits the aggregation of human platelets (24).
Previously, we found that treatment of mES cells with various HDAC inhibitors induced a decrease of stem cell marker expression and morphologically-identified differentiation of mES cells (25). To investigate the effects of the HDAC inhibitors on the differentiation of specific lineages, we performed hematopoietic differentiation experiments using several HDAC inhibitors such as sodium butyrate, valproic acid, and nicotinamide and monitored the expression patterns of key differentiation-related genes using RT-PCR. Among the HDAC inhibitors we tested, nicotinamide enhanced the expression of mesodermal marker genes such as Scl and Flk1. To further evaluate the implications of the data, we here investigated the effects of nicotinamide and splitomicin on the hematopoietic differentiation of mES cells in detail and found that the modulation of the SIRT1 activity can enhance the yield of hematopoietic differentiation to specific lineages.
The mES cell line R1 was maintained on mouse embryonic fibroblast (MEF) cells in ES medium, which contains Dulbecco’s modified Eagle’s medium (DMEM, Hyclone Inc., Logan, UT, USA), 15% fetal bovine serum (FBS, Hyclone Inc.), 0.1 mM β-mercaptoethanol (Sigma Aldrich, St. Louis, MO, USA), 0.1 mM nonessential amino acids (Invitrogen, Carlsbad, CA, USA), 100 U/ml penicillin and 100 μg/ml streptomycin (Hyclone Inc.), at 37°C under a humidified atmosphere of 5% CO2. ES cells were dissociated with TrypLETM Express (Gibco, Invitrogen, Carlsbad, CA, USA) and then seeded on MEF cells prepared as follows. MEF cells were harvested and irradiated with 50 grays and seeded at a density of ~5.5×104 cells/ml in MEF medium (DMEM, 10% FBS, 0.1 mM β-mercaptoethanol, 0.1 mM nonessential amino acids, 100 U/ml penicillin and 100 μg/ml streptomycin) a day before ES cell seeding.
Differentiation of mES cells to a hematopoietic lineage based on a semi-solid culture system was accomplished using protocols obtained from Stem Cell Technologies (Vancouver, British Columbia, Canada). For the primary differentiation (EB formation), mES cells were trypsinized into a single cell suspension and re-suspended in the primary differentiation medium (Iscove’s Modified Dulbecco’s Medium (IMDM, Hyclone Inc.), 1% methylcellulose (Methocult M3120, Stem Cell Technologies), 15% FBS, 2 mM L-Glutamine (Sigma Aldrich), 150 μM monothioglycerol (MTG, Sigma Aldrich) and 40 ng/ml mouse SCF (PeproTech, Rocky Hill, NJ, USA)) at a density of 2×103 cells/ml. The cell suspension (1 ml) was plated onto a 35 mm dish and incubated at 37°C with 5% CO2 for 10 days. On day 7, to ensure the viability of the primary differentiation cultures over an extended period of time, feed medium (IMDM, 0.5% methylcellulose, 15% FBS, 150 μM MTG, 160 ng/ml mouse SCF, 30 ng/ml mouse IL-3 (PeproTech), 20 ng/ml human IL-6 (Genscript, Scotch Plains, NJ, USA), and 3 U/ml human EPO (R&D Systems, Minneapolis, MN, USA)) was added. For the secondary differentiation (clonal assays of EBs), EBs were digested with TrypLETM Express followed by passaging through a 21 gauge needle 3 to 5 times and plated at a density of 1×104 cells/ml with hematopoietic differentiation medium (IMDM, 1% methylcellulose, 15% FBS, 2 mM L-Glutamine, 150 μM MTG, BIT 9500 Serum Substitute (1% BSA, 10 μg/ml Insulin, 200 μg/ml Transferrin; Stem Cell Technologies), 150 ng/ml mouse SCF, 30 ng/ml mouse IL-3, 30 ng/ml human IL-6, and 3 U/ml human EPO) with or without SIRT1 inhibitors (nicotinamide or splitomicin) onto 35 mm dishes and incubated at 37°C with 5% CO2. After 7 days, the colonies were identified and counted. Individual colonies were either identified by the morphological characteristics of the different kinds of hematopoietic cell types or harvested from the methylcellulose medium for PCR and flow cytometry analyses. SIRT1 inhibitors were purchased from Sigma-Aldrich.
Hematopoietic colonies were trypsinized into a single cell suspension and washed with 5% FBS in phosphate buffered saline (PBS). The total cell numbers were determined and adjusted to a concentration of 0.5~1×106 cells/ml in ice cold PBS. The cells were preincubated with Rat Anti-Mouse CD16/CD32 antibody for 10 minutes at 4°C, incubated with PE-conjugated primary antibodies (Rat Anti-Mouse CD11b, Rat Anti-Mouse Ly-6G and Ly-6C (Gr-1), Rat Anti-Mouse TER-119, Rat Anti-Mouse CD45, Rat Anti-Mouse CD41 and Rat Anti-Mouse CD117) for 40 minutes at 4°C, washed twice, and analyzed by FACSCalibur flow cytometry (BD Biosciences, Bedford, MA, USA). All antibodies were purchased from BD Biosciences. The data were analyzed with the software FCS Express 5 (De Novo Software, Los Angeles, CA, USA).
Total RNAs were isolated using TRI Reagent® according to the manufacturer’s instructions (MRC, Cincinnati, OH, USA). Then, 2 μg of the total RNA was reverse-transcribed in the first-strand buffer containing 6 μg/ml oligo (dT) primer, 50 U M-MLV reverse transcriptase (Invitrogen), 2 mM dNTP, and 40 U RNaseOUT recombinant ribonuclease inhibitor (Invitrogen). The reaction was conducted at 42°C for 1 hour. One microliter of the cDNA synthesis was subjected to a standard PCR reaction of 20~35 cycles of denaturation for 60 seconds at 95°C, annealing for 60 seconds at 58~60°C, and elongation for 60 seconds at 72°C. The primer sequences used were as follows: Brachyury, forward 5′-TGCTGCCTGTGAGTCATAAC-3′ and reverse 5′-TCCAGGTGCTATATATTGCC-3′; Flk1, forward 5′-TAGGTGCCTCCCCATACCCTGG-3′ and reverse 5′-TGGCCGGCTCTTTCGCTTACTG-3′; Scl, forward 5′-TATGAGATGGAGATTTCTGATG-3′ and reverse 5′-GCTCCTCTGTGTAACTGTCC-3′; GATA1, forward 5′-GGAATTCGGGCCCCTTGTGAGGCCAGAGAG-3′ and reverse 5′-CGGGGTACCTCACGCTCCAGCCAGATTCGA-3′; GATA2, forward 5′-CTCCAGCTTCACCCCTAAGCAG-3′ and reverse 5′-CATAAGGTGGTGGTTGTCGTCT-3′; βH1, forward 5′-AGTCCCCATGGAGTCAAAGA-3′ and reverse 5′-CTCAAGGAGACCTTTGCTCA-3′; β-major, forward 5′-CTGACAGATGCTCTCTTGGG-3′ and reverse 5′-CACAAACCCCAGAAACAGACA-3′; Pu.1, forward 5′-TGGAAGGGTTTTCCCTCACC-3′ and reverse 5′-TGCTGTCCTTCATGTCGCCG-3′; HoxB4, forward 5′-CTGGATGCGCAAAGTTCAC-3′ and reverse 5′-GTGAAACTCCTTCTCCAACTCC-3′; IL-7R, forward 5′-TTACTTCAAAGGCTTCTGGAG-3′ and reverse 5′-CTGGCTTCAACGCCTTTCACCTCA-3′; Rag2, forward 5′-ATTCCTGGCTTGGCCGAAAGG-3′ and reverse 5′-TAGGACAGGATCTCTTAGGCC-3′; GATA3, forward 5′-TATGTGCCCGAGTACAGCTC-3′ and reverse 5′-TGTAGTACAGCCCACAGGCA-3′; and GAPDH, forward 5′-ACCACAGTCCATGCCATCAC-3′ and reverse 5′-TCCACCACCCTGTTGCTGTA-3′.
To test the possible effect of the SIRT1 inhibitors on hematopoietic differentiation in detail, we used a two-step in vitro hematopoietic differentiation protocol. In the first step, mES cells were suspended as single cells in a methylcellulose-based medium and cultured for 10 days which promotes primary differentiation. In the second step, EBs were dissociated into single cells and re-plated in methylcellulose-based medium containing a cocktail of cytokines (SCF, IL-3, IL-6, and EPO) to examine their ability to form hematopoietic colonies. At this stage, the cells were simultaneously treated with or without SIRT1 inhibitors and cultured for 21 days (Fig. 1).
We counted the hematopoietic colonies on day 7 from secondary differentiation and evaluated the effects of SIRT1 inhibition on hematopoietic cell growth and progenitor differentiation. EB-derived cells, which were differentiated with or without the SIRT1 inhibitor, had a similar potential of hematopoietic development in terms of the differentiation lineages: BFU-E (burst-forming unit erythroid), CFU-E (colony-forming unit erythroid), CFU-GM (colony-forming unit granulocyte, macrophage), CFU-GEMM (colony-forming unit-granulocyte, erythrocyte, monocyte/macrophage, and megakaryocyte) and 2nd EB (secondary EB) were commonly formed in both conditions on day 7 of the culture (Fig. 2A). Secondary EBs are thought to be derived from the cells of primary EBs, and they have not yet undergone lineage commitment and subsequent differentiation. When we quantified the colonies of different lineages, the SIRT1 inhibitor-treated groups had higher total numbers of colonies compared with the control group on day 7: the total numbers of colonies were 140.8±11.96 (control), 183.2±15.02 (nicotinamide-treated group) and 159.5± 6.60 (splitomicin-treated group), respectively (Table 1). Especially, the total number of colonies in the nicotinamide-treated group was significantly higher than that in the control group (Fig. 2B).
As shown in Fig. 2B, the higher numbers of hematopoietic progenitor colonies, BFU-E, CFU-E and CFU-GM, were generated from the SIRT1 inhibitor-treated groups compared with the control group. The number of BFU-E (12.8±3.67 vs. 9.5±3.36), CFU-E (65.9±7.80 vs. 40.9±4.59) and CFU-GM (43.9±6.57 vs. 31.5±4.45) colonies significantly increased in the nicotinamide-treated group compared to the control group. The treatment of splitomicin significantly increased the number of BFU-E (14.4±3.50 vs. 9.5±3.36) and CFU-GM (44.4±5.24 vs. 31.5±4.45) colonies compared to the control group. In addition, the percentage of BFU-E, CFU-E and CFU-GM colonies tended to increase in the groups treated with nicotinamide and splitomicin (Fig. 2C). These results suggest that the SIRT1 inhibitors may influence the formation and/or differentiation of hematopoietic progenitors.
We next investigated whether the treatment of the SIRT1 inhibitors altered the expression of mesodermal and hematopoietic lineage markers. We selected the Brachyury, Flk1, Scl, GATA1, GATA2, HoxB4, βH1, β-major and PU.1 as marker genes. We analyzed the expression of these genes using RT-PCR with the colonies obtained on days 7, 14, and 21 of second differentiation (Fig. 3A). Expression of Brachyury, Flk1, Scl, HoxB4 and GATA2 mRNA did not change in the presence of the SIRT1 inhibitors. However, expression of GATA1, βH1, β-major globin and Pu.1 were upregulated in the nicotinamide-and splitomicin-treated group compared to the control group. This result is associated with the enhanced numbers of CFU-E, BFU-E and CFU-GM colonies in the presence of the SIRT1 inhibitors (Fig. 2B). To further test the effect of SIRT1 inhibitors on lymphoid differentiation from ES-derived EBs, we analyzed the expression of lymphoid lineage marker genes such as IL-7R, Rag2 and GATA3 using RT-PCR. As shown Fig. 3B, SIRT1 inhibitors during hematopoietic differentiation didn’t change lymphoid lineage marker gene expression compared to the control group. Taken together, these results suggest that the inhibition of SIRT1 during the secondary differentiation process may induce increased production of erythroid and myeloid lineage cells.
To directly determine whether the treatment with the SIRT1 inhibitors during the hematopoietic differentiation increases the fate determination of erythroid and myeloid lineage cells, we analyzed the blood cell population in the colonies obtained on days 7 and 14 of secondary differentiation based on the expression level of hematopoietic cell-specific surface proteins measured by flow cytometry (Fig. 4).
CD117 is expressed on hematopoietic progenitor cells, lymphoid, myeloid, erythroid, and megakaryocyte progenitors (26). CD41 expression is associated with the earliest stages of mouse hematopoiesis and recognized as a megakaryocyte/platelet marker (27). Therefore, CD117 and CD41 were used as surface markers for hematopoietic stem cells and progenitor cells. The change in the population of CD117 positive cells and CD41 positive cells was not significant among the tested groups.
CD45 is a commonly used pan-hematopoietic lineage marker (28). TER-119, an erythroid lineage marker, is expressed on erythroid cells from early pro-erythroblast to mature erythrocyte stages (29). CD11b is a commonly used marker for the monocyte/macrophage lineage cells (30). Gr-1 has been used successfully to separate mouse granulocyte lineage cells (31). As shown in Fig. 4A, nicotinamide slightly increased the percentage of Ter-119 positive cells but decreased Gr-1 positive cell population on day 7 compared to the control group. On the other hand, the percentage of Ter-119 positive cells decreased on day 14 after treatment with nicotinamide (Fig. 4B). There was no change in expression of other makers on days 7 and 14. However, the treatment with splitomicin increased percentage of CD45, TER-119, CD11b, and Gr-1 positive cells compared to the control group on day 7 (Fig. 4A). In addition, the treatment of splitomicin consistently increased the percentage of Ter-119, CD11b, and Gr-1 positive cells on day 14 (Fig. 4B). These results suggest that splitomicin may improve the generation of myeloid lineage cells including erythroid cells, monocytes/macrophages and granulocytes at the differentiation stage from EBs to hematopoietic progenitor cells.
Hematopoietic stem cells (HSCs) are one of the most studied and characterized stem cells. HSCs can be isolated from blood and bone marrow and can self-renew and differentiate into a variety of blood cells which could be used to treat diverse hematological diseases (32). Hematopoietic cells are also considered as a typical model system to understand the mechanism of ES cell differentiation. In this study, we report that the treatment with SIRT1 inhibitors in hematopoietic differentiation from ES cells increase the yield of myeloid and erythroid cell lineages.
During the past several decades, many researchers have been trying to get more numbers of HSCs and hematopoietic progenitor cells (HPCs) from ES cells. Despite the extensive studies, hematopoietic cell derivation from ES cells is still difficult. The developed differentiation method using ES cells has low efficiency and requires many procedures, costs, and time. Genetic manipulation of mES cells was used to improve understanding of HSC generation and differentiation. For example, a homeobox transcription factor, HoxB4, is a key factor in switching EB-derived hematopoietic progenitors into definitive HSCs (33). Ectopic overexpression of HoxB4 in mES cells can enhance self-renewal and expansion of hematopoietic progenitor cells differentiated from mES cells (34). The PcG protein Bmi1 is an essential regulator in adult HSCs. Ectopic expression of Bmi1 enhanced the development of hematopoietic cells during ES cell differentiation (35). However, ES cells generated by genetic manipulation may be more difficult when considering clinical applications. Therefore, application of small molecules such as SIRT1 inhibitors in hematopoietic differentiation may be helpful for a more efficient generation of hematopoietic cells.
SIRT1 modulates pluripotency, differentiation and stress response in mouse ES cells (36). SIRT1 is highly expressed in mES cells and downregulated during differentiation processes (37). Saunders et al. showed that inhibition of SIRT1 protein expression by miRNAs can have an important role in development because several miRNAs targeting SIRT1 have been identified as regulators of a specific differentiation pathway (38). Previous studies reported that SIRT1 is essential for hematopoietic lineage specification and homeostatic maintenance of HSCs. SIRT1 knockout ES cells formed fewer mature blast cell colonies, and SIRT1 knockout yolk sacs exhibited fewer primitive erythroid precursors (17). The fetal liver hematopoietic progenitors isolated from a SIRT1 knockout mouse also had a significantly lower hematopoietic potential compared to normal progenitors (39). These results show that sustained SIRT1 inhibition has a negative effect on hematopoietic differentiation and lineage commitment. However, when bone marrow cells derived from a SIRT1 knockout mouse were transplanted into irradiated animals, there was no prominent defect in the context of the HSC properties and function such as production of mature blood cells, lineage distribution and frequencies of primitive HSC populations (40). In our experiments, we formed EBs first and then added SIRT1 inhibitors during the secondary hematopoietic differentiation. In this condition, higher numbers of hematopoietic colony forming units were generated suggesting that the SIRT1 inhibitors may positively affect the formation of hematopoietic progenitor cells differentiated from EBs. Treatment with SIRT1 inhibitors also enhanced the expression of primitive and definitive hematopoietic specific genes such as GATA1, β H1, Pu.1, and β-major. However, cell population analysis revealed that among the SIRT1 inhibitors, only treatment with splitomicin can consistently increase the population of erythroid and myeloid lineage cells. Although we can’t explain specific mechanisms involved in the complex effects of nicotinamide, these results suggest that the treatment of the SIRT1 inhibitors can modulate the hematopoietic differentiation of mouse ES cells, and transient treatment of splitomicin at some stage of differentiation into hematopoietic lineage may improve hematopoietic progenitor production. Therefore, we suggest that an optimized treatment with SIRT1 inhibitors in the hematopoietic differentiation of mES cells may provide a better protocol to obtain a hematopoietic lineage of cells.
Acknowledgments
This research was supported by a grant from the National Research Foundation (2017M3A9B4065302) funded by the Ministry of Science, ICT and Future Planning in the Republic of Korea.
References
1. Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998; 282:1145–1147. DOI: 10.1126/science.282.5391.1145. PMID: 9804556.

2. Boiani M, Schöler HR. Regulatory networks in embryo-derived pluripotent stem cells. Nat Rev Mol Cell Biol. 2005; 6:872–884. DOI: 10.1038/nrm1744. PMID: 16227977.

3. Kaufman DS. Toward clinical therapies using hematopoietic cells derived from human pluripotent stem cells. Blood. 2009; 114:3513–3523. DOI: 10.1182/blood-2009-03-191304. PMID: 19652198. PMCID: 2766673.

4. Keller GM. In vitro differentiation of embryonic stem cells. Curr Opin Cell Biol. 1995; 7:862–869. DOI: 10.1016/0955-0674(95)80071-9. PMID: 8608017.

5. Keller G, Kennedy M, Papayannopoulou T, Wiles MV. Hematopoietic commitment during embryonic stem cell differentiation in culture. Mol Cell Biol. 1993; 13:473–486. DOI: 10.1128/MCB.13.1.473. PMID: 8417345. PMCID: 358927.

6. Yamane T, Hosen N, Yamazaki H, Weissman IL. Expression of AA4.1 marks lymphohematopoietic progenitors in early mouse development. Proc Natl Acad Sci U S A. 2009; 106:8953–8958. DOI: 10.1073/pnas.0904090106. PMID: 19458045. PMCID: 2690003.

7. Wiles MV, Keller G. Multiple hematopoietic lineages develop from embryonic stem (ES) cells in culture. Development. 1991; 111:259–267. PMID: 1893864.

8. Chadwick K, Wang L, Li L, Menendez P, Murdoch B, Rouleau A, Bhatia M. Cytokines and BMP-4 promote hematopoietic differentiation of human embryonic stem cells. Blood. 2003; 102:906–915. DOI: 10.1182/blood-2003-03-0832. PMID: 12702499.

9. Herrmann BG. Expression pattern of the Brachyury gene in whole-mount TWis/TWis mutant embryos. Development. 1991; 113:913–917. PMID: 1821859.

10. Shalaby F, Rossant J, Yamaguchi TP, Gertsenstein M, Wu XF, Breitman ML, Schuh AC. Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature. 1995; 376:62–66. DOI: 10.1038/376062a0. PMID: 7596435.

11. Porcher C, Swat W, Rockwell K, Fujiwara Y, Alt FW, Orkin SH. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell. 1996; 86:47–57. DOI: 10.1016/S0092-8674(00)80076-8. PMID: 8689686.

12. Orkin SH. GATA-binding transcription factors in hematopoietic cells. Blood. 1992; 80:575–581. PMID: 1638017.

13. Nerlov C, Graf T. PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev. 1998; 12:2403–2412. DOI: 10.1101/gad.12.15.2403. PMID: 9694804. PMCID: 317050.

14. Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007; 404:1–13. DOI: 10.1042/BJ20070140. PMID: 17447894. PMCID: 2753453.

15. Sakamoto J, Miura T, Shimamoto K, Horio Y. Predominant expression of Sir2alpha, an NAD-dependent histone deacetylase, in the embryonic mouse heart and brain. FEBS Lett. 2004; 556:281–286. DOI: 10.1016/S0014-5793(03)01444-3. PMID: 14706864.

16. Homma K, Sone M, Taura D, Yamahara K, Suzuki Y, Takahashi K, Sonoyama T, Inuzuka M, Fukunaga Y, Tamura N, Itoh H, Yamanaka S, Nakao K. Sirt1 plays an important role in mediating greater functionality of human ES/iPS-derived vascular endothelial cells. Atherosclerosis. 2010; 212:42–47. DOI: 10.1016/j.atherosclerosis.2010.04.021. PMID: 20488443.

17. Ou X, Chae HD, Wang RH, Shelley WC, Cooper S, Taylor T, Kim YJ, Deng CX, Yoder MC, Broxmeyer HE. SIRT1 deficiency compromises mouse embryonic stem cell hematopoietic differentiation, and embryonic and adult hematopoiesis in the mouse. Blood. 2011; 117:440–450. DOI: 10.1182/blood-2010-03-273011. PMID: 20966168. PMCID: 3031475.

18. Berger F, Ramírez-Hernández MH, Ziegler M. The new life of a centenarian: signalling functions of NAD(P). Trends Biochem Sci. 2004; 29:111–118. DOI: 10.1016/j.tibs.2004.01.007. PMID: 15003268.

19. Denu JM. Vitamin B3 and sirtuin function. Trends Biochem Sci. 2005; 30:479–483. DOI: 10.1016/j.tibs.2005.07.004. PMID: 16039130.

20. Giammona LM, Fuhrken PG, Papoutsakis ET, Miller WM. Nicotinamide (vitamin B3) increases the polyploidisation and proplatelet formation of cultured primary human megakaryocytes. Br J Haematol. 2006; 135:554–566. DOI: 10.1111/j.1365-2141.2006.06341.x. PMID: 17054670.

21. Peled T, Shoham H, Aschengrau D, Yackoubov D, Frei G, Rosenheimer GN, Lerrer B, Cohen HY, Nagler A, Fibach E, Peled A. Nicotinamide, a SIRT1 inhibitor, inhibits differentiation and facilitates expansion of hematopoietic progenitor cells with enhanced bone marrow homing and engraftment. Exp Hematol. 2012; 40:342–355.e1. DOI: 10.1016/j.exphem.2011.12.005. PMID: 22198152.
22. Posakony J, Hirao M, Stevens S, Simon JA, Bedalov A. Inhibitors of Sir2: evaluation of splitomicin analogues. J Med Chem. 2004; 47:2635–2644. DOI: 10.1021/jm030473r. PMID: 15115404.

23. Neugebauer RC, Uchiechowska U, Meier R, Hruby H, Valkov V, Verdin E, Sippl W, Jung M. Structure-activity studies on splitomicin derivatives as sirtuin inhibitors and computational prediction of binding mode. J Med Chem. 2008; 51:1203–1213. DOI: 10.1021/jm700972e. PMID: 18269226.

24. Liu FC, Liao CH, Chang YW, Liou JT, Day YJ. Splitomicin suppresses human platelet aggregation via inhibition of cyclic AMP phosphodiesterase and intracellular Ca++ release. Thromb Res. 2009; 124:199–207. DOI: 10.1016/j.thromres.2009.02.013. PMID: 19327818.

25. Park JA, Kim YE, Seok HJ, Park WY, Kwon HJ, Lee Y. Differentiation and upregulation of heat shock protein 70 induced by a subset of histone deacetylase inhibitors in mouse and human embryonic stem cells. BMB Rep. 2011; 44:176–181. DOI: 10.5483/BMBRep.2011.44.3.176. PMID: 21429295.

26. Ogawa M, Matsuzaki Y, Nishikawa S, Hayashi S, Kunisada T, Sudo T, Kina T, Nakauchi H, Nishikawa S. Expression and function of c-kit in hemopoietic progenitor cells. J Exp Med. 1991; 174:63–71. DOI: 10.1084/jem.174.1.63. PMID: 1711568. PMCID: 2118893.

27. Robin C, Ottersbach K, Boisset JC, Oziemlak A, Dzierzak E. CD41 is developmentally regulated and differentially expressed on mouse hematopoietic stem cells. Blood. 2011; 117:5088–5091. DOI: 10.1182/blood-2011-01-329516. PMID: 21415271. PMCID: 3109535.

28. McKinney-Freeman SL, Naveiras O, Yates F, Loewer S, Philitas M, Curran M, Park PJ, Daley GQ. Surface antigen phenotypes of hematopoietic stem cells from embryos and murine embryonic stem cells. Blood. 2009; 114:268–278. DOI: 10.1182/blood-2008-12-193888. PMID: 19420357. PMCID: 2714203.

29. Kina T, Ikuta K, Takayama E, Wada K, Majumdar AS, Weissman IL, Katsura Y. The monoclonal antibody TER-119 recognizes a molecule associated with glycophorin A and specifically marks the late stages of murine erythroid lineage. Br J Haematol. 2000; 109:280–287. DOI: 10.1046/j.1365-2141.2000.02037.x. PMID: 10848813.

30. Lai L, Alaverdi N, Maltais L, Morse HC 3rd. Mouse cell surface antigens: nomenclature and immunophenotyping. J Immunol. 1998; 160:3861–3868. PMID: 9558091.
31. Fleming TJ, Fleming ML, Malek TR. Selective expression of Ly-6G on myeloid lineage cells in mouse bone marrow. RB6–8C5 mAb to granulocyte-differentiation antigen (Gr-1) detects members of the Ly-6 family. J Immunol. 1993; 151:2399–2408. PMID: 8360469.
32. Avior Y, Sagi I, Benvenisty N. Pluripotent stem cells in disease modelling and drug discovery. Nat Rev Mol Cell Biol. 2016; 17:170–182. DOI: 10.1038/nrm.2015.27. PMID: 26818440.

33. Kyba M, Perlingeiro RC, Daley GQ. HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell. 2002; 109:29–37. DOI: 10.1016/S0092-8674(02)00680-3. PMID: 11955444.

34. Jackson M, Ma R, Taylor AH, Axton RA, Easterbrook J, Kydonaki M, Olivier E, Marenah L, Stanley EG, Elefanty AG, Mountford JC, Forrester LM. Enforced expression of HOXB4 in human embryonic stem cells enhances the production of hematopoietic progenitors but has no effect on the maturation of red blood cells. Stem Cells Transl Med. 2016; 5:981–990. DOI: 10.5966/sctm.2015-0324. PMID: 27352929. PMCID: 4954454.

35. Ding X, Lin Q, Ensenat-Waser R, Rose-John S, Zenke M. Polycomb group protein Bmi1 promotes hematopoietic cell development from embryonic stem cells. Stem Cells Dev. 2012; 21:121–132. DOI: 10.1089/scd.2010.0539. PMID: 21545235.

36. Han MK, Song EK, Guo Y, Ou X, Mantel C, Broxmeyer HE. SIRT1 regulates apoptosis and Nanog expression in mouse embryonic stem cells by controlling p53 subcellular localization. Cell Stem Cell. 2008; 2:241–251. DOI: 10.1016/j.stem.2008.01.002. PMID: 18371449. PMCID: 2819008.

37. Calvanese V, Lara E, Suárez-Alvarez B, Abu Dawud R, Vázquez-Chantada M, Martínez-Chantar ML, Embade N, López-Nieva P, Horrillo A, Hmadcha A, Soria B, Piazzolla D, Herranz D, Serrano M, Mato JM, Andrews PW, López-Larrea C, Esteller M, Fraga MF. Sirtuin 1 regulation of developmental genes during differentiation of stem cells. Proc Natl Acad Sci U S A. 2010; 107:13736–13741. DOI: 10.1073/pnas.1001399107. PMID: 20631301. PMCID: 2922228.

38. Saunders LR, Sharma AD, Tawney J, Nakagawa M, Okita K, Yamanaka S, Willenbring H, Verdin E. miRNAs regulate SIRT1 expression during mouse embryonic stem cell differentiation and in adult mouse tissues. Aging (Albany NY). 2010; 2:415–431. DOI: 10.18632/aging.100176. PMID: 20634564. PMCID: 2933889.

39. Matsui K, Ezoe S, Oritani K, Shibata M, Tokunaga M, Fujita N, Tanimura A, Sudo T, Tanaka H, McBurney MW, Matsumura I, Kanakura Y. NAD-dependent histone deacetylase, SIRT1, plays essential roles in the maintenance of hematopoietic stem cells. Biochem Biophys Res Commun. 2012; 418:811–817. DOI: 10.1016/j.bbrc.2012.01.109. PMID: 22306819.

40. Leko V, Varnum-Finney B, Li H, Gu Y, Flowers D, Nourigat C, Bernstein ID, Bedalov A. SIRT1 is dispensable for function of hematopoietic stem cells in adult mice. Blood. 2012; 119:1856–1860. DOI: 10.1182/blood-2011-09-377077. PMID: 22219225. PMCID: 3293640.

Fig. 1
Schematic representation of the culture system used for hematopoietic cell differentiation from mouse ES cells. For hematopoietic EB formation, ES cells were differentiated with the methylcellulose medium with SCF for 10 days. For secondary differentiation, EBs were harvested and disrupted into single cells and replated with cytokines (SCF, IL-3, IL-6, and EPO) in the presence or absence of SIRT1 inhibitors. Counting of the colony numbers, RT-PCR and FACS analyses were performed at the indicated time points.
Fig. 2
SIRT1 inhibitors enhance hematopoietic colony forming ability during hematopoietic differentiation of mES cells. (A) Colony-forming potential of mES-derived cells in the presence of nicotinamide 0.5 mM (NAM) and splitomicin 60 μM (SP). ES cells were differentiated by EB formation for 10 days. EBs were disrupted into single cells and re-plated in 1% methylcellulose medium containing a cocktail of hematopoietic cytokines. Phase-contrast images were taken from representative BFU-E, CFU-E, CFU-GM and CFU-GEMM colonies after 7 days. Scale bar. 100 μm. (B) After the colony-forming assays, the numbers of BFU, CFU-E, CFU-GM, CFU-GEMM, and 2nd EB were scored by microscopy and compared. Each bar is expressed as the mean±standard error of the mean. *p<0.05, **p<0.01, ***p<0.001 (vs. control) of n=9 independent experiments. (C) The results are shown in percentage terms. Percentages represent the proportion of each colony type.
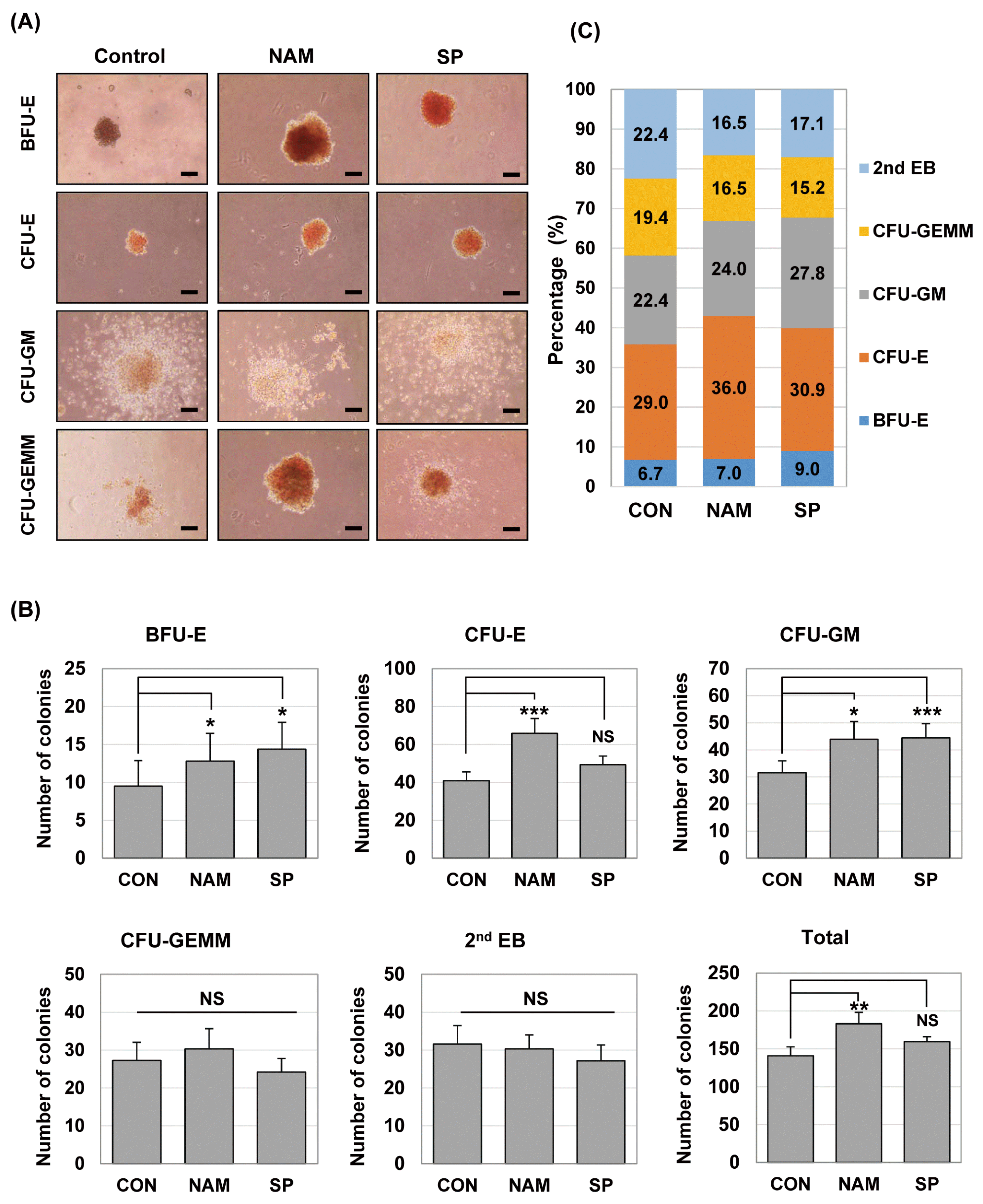
Fig. 3
Effects of SIRT1 inhibitors on the expression of mesodermal and hematopoietic marker genes during the hematopoietic differentiation of mES cells. (A) RNA samples were extracted on days 7, 14, and 21 during second hematopoietic differentiation, and gene expression was analyzed by RT-PCR. GAPDH is shown as a control. ES: mES cells, EB 10D: embryoid bodies obtained on day 10 of primary differentiation. Mesodermal marker, brachyury; hematopoietic stem cell and progenitor cell markers, Scl, Flk1, and HoxB4; hematopoietic lineage cell markers, GATA1 and GATA2; erythroid specific markers, βH-1 and β-major; early myeloid and lymphoid progenitor marker, Pu.1. The density of each band was measured using the ImageJ program and normalized with the density of GAPDH used as a loading control for each sample. The relative band density values were compared with the controls on day 7 (shown as fold expression). (B) RNA samples were extracted on days 7 and 14 during second hematopoietic differentiation, and gene expression was analyzed by RT-PCR. IL-7R, Rag2, and GATA3 were used as lymphoid lineage markers.
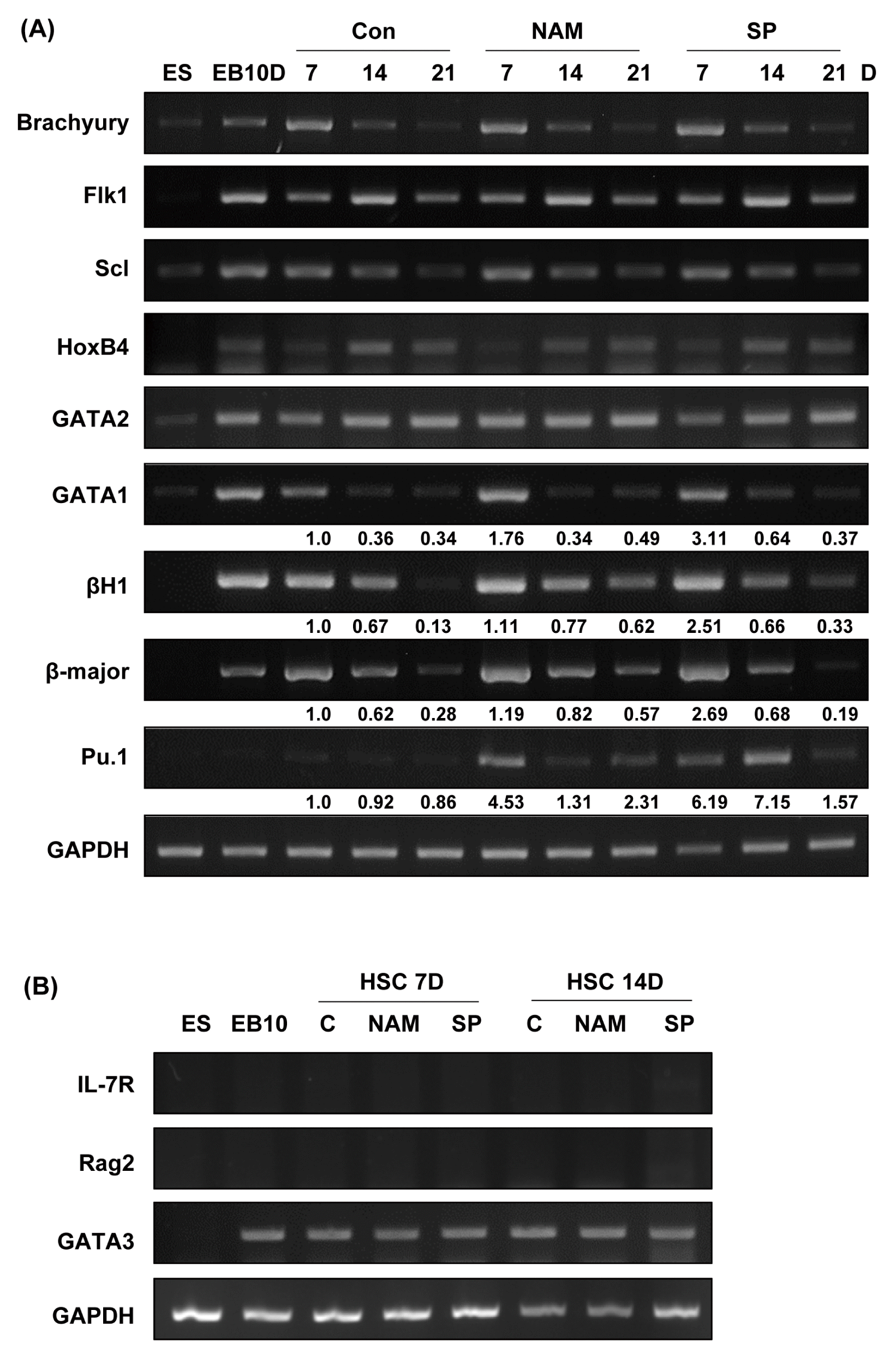
Fig. 4
Effects of SIRT1 inhibitors on the expression of hematopoietic surface antigens during hematopoietic differentiation. Expression of surface antigens was analyzed by flow cytometry. (A) Expression of CD41 and CD117 (hematopoietic progenitors), CD45 (hematopoietic), TER-119 (erythroid), CD11b (monocyte/macrophage) and Gr-1 (granulocyte) markers was monitored on day 7 of secondary hematopoietic differentiation. (B) Expression of TER-119, CD11b and Gr-1 markers was monitored on day 14 of secondary hematopoietic differentiation. EB 10D, embryoid bodies obtained on day 10 of primary differentiation. Each bar is expressed as the mean±standard error of the mean of three experiments. *p<0.05, **p<0.01 (vs. control).
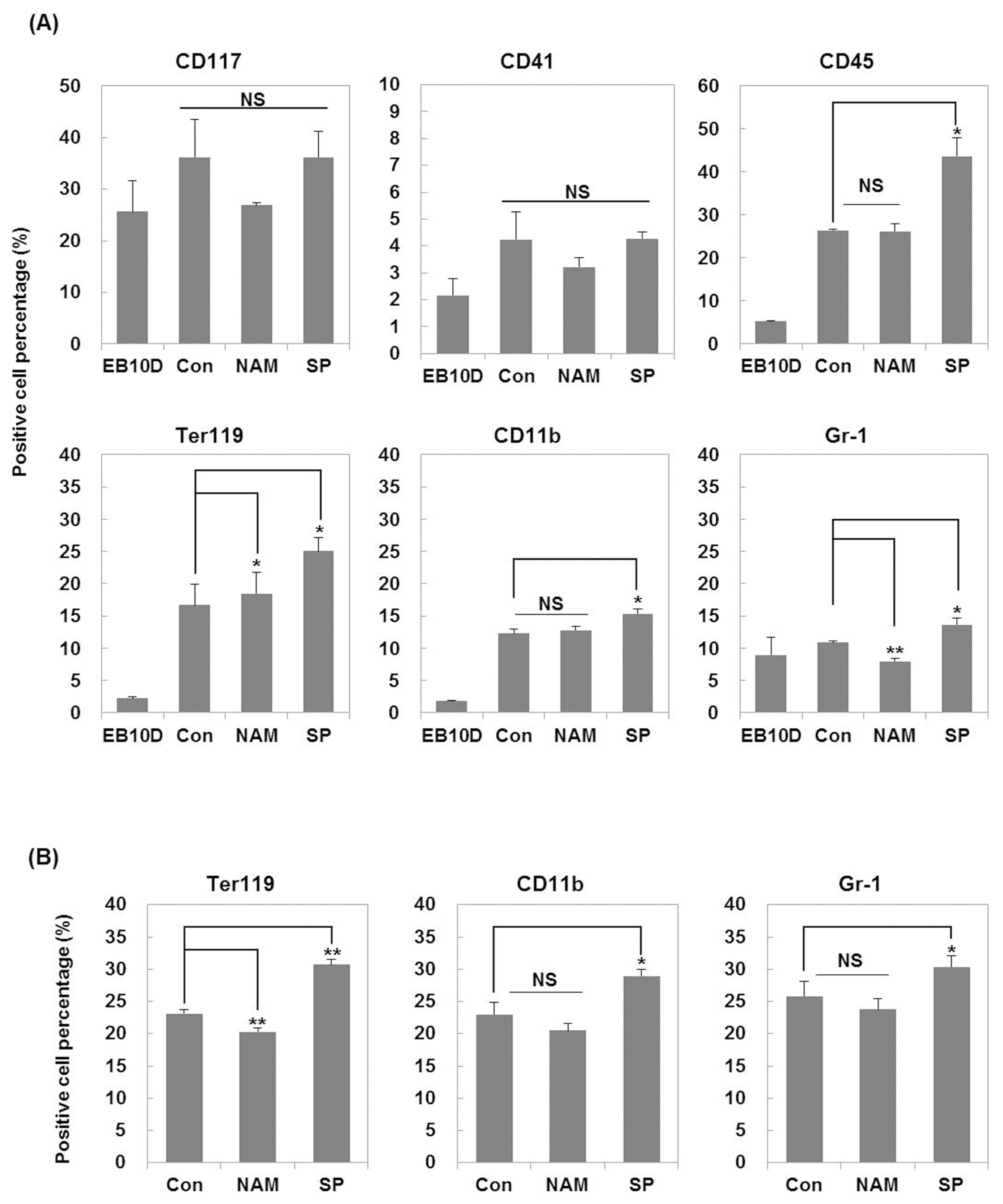
Table 1
The effect of the SIRT1 inhibitors on the numbers of mES cell-derived hematopoietic colonies
The total numbers of BFU, CFU-E, CFU-GM, CFU-GEMM, and 2nd EB in each group were scored by microscopy on day 7. Values are expressed as the mean±standard error of the mean per 104 cells (n=9). BFU-E: burst forming unit-erythroid, CFU-E: colony forming units-erythroid, CFU-GM: colony forming unit-granulocyte, macrophage, CFU-GEMM: colony forming unit-granulocyte, erythrocyte, macrophage, megakaryocyte, 2nd EB: secondary embryoid body.




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download