Abstract
Improved approaches for promoting umbilical cord blood (CB) hematopoietic stem cell (HSC) homing are clinically important to enhance engraftment of CB-HSCs. Clinical transplantation of CB-HSCs is used to treat a wide range of disorders. However, an improved understanding of HSC chemotaxis is needed for facilitation of the engraftment process. We found that ectopic overexpression of miR-9 and antisense-miR-9 respectively down- and up-regulated C-X-C chemokine receptor type 4 (CXCR4) expression in CB-CD34+ cells as well as in 293T and TF-1 cell lines. Since CXCR4 is a specific receptor for the stromal cell derived factor-1 (SDF-1) chemotactic factor, we investigated whether sense miR-9 and antisense miR-9 influenced CXCR4-mediated chemotactic mobility of primary CB CD34+ cells and TF-1 cells. Ectopic overexpression of sense miR-9 and antisense miR-9 respectively down- and up-regulated SDF-1-mediated chemotactic cell mobility. To our knowledge, this study is the first to report that miR-9 may play a role in regulating CXCR4 expression and SDF-1-mediated chemotactic activity of CB CD34+ cells.
CXCR4 and the chemokine SDF-1, also known as C-X-C motif chemokine 12 (CXCL12), are involved in the migration of HSCs. CXCR4 is expressed in immature hematopoietic cells, and stromal cells of the bone marrow (BM) are a main source of SDF-1 (1). CXCR4 expression levels in CD34+ cells, a highly enriched population of HSC and hematopoietic progenitor cells, were correlated to repopulation rates following allogeneic CB hematopoietic cell transplantation (HCT). CB-derived CD34+ cells treated with an antibody to CXCR4 display reduced engraftment compared to those cells treated with control antibody (2, 3). In addition, intravenously injected human SDF-1 increases the homing of human mobilized peripheral blood- and CB-CD34+-enriched cells to the BM and spleen of NOD/SCID mice (4, 5). Mice lacking SDF-1 or CXCR4 exhibit many defects, including the absence of both lymphoid and myeloid hematopoiesis in fetal BM (6, 7). Conversely, ectopic expression of CXCR4 in the HSCs leads to better response to SDF-1 and more efficient engraftment (8).
CB is a clinical source of HSCs used in transplantations related to the treatment of various malignant and non-malignant diseases (9, 10). However, there are disadvantages associated with the use of CB HSCs for HCT, compared to the use of cells from BM and mobilized peripheral blood. One disadvantage, compared to other HSC sources, is slow neutrophil, platelet, and immune cell reconstitution (9, 10). Recent efforts to improve CB-HCT have focused on increasing CXCR4 expression in HSCs to enhance their migration to the BM (11, 12). However, the process by which microRNAs (miRNAs) may influence CXCR4 expression in CD34+ CB cells remains unexplored.
miRNAs are short, noncoding RNAs containing approximately 22 nucleotides. Studies indicate that they suppress gene expression or play a role in gene cleavage through sequence-specific base pairing with target mRNAs to form an RNA-induced silencing complex (13–16).
miRNAs suppress a set of functionally related genes and are thus involved in most cell biological processes, including cell viability, proliferation, development, and differentiation (i.e. HSC differentiation). In particular, a specific miRNA expression pattern occurs at the HSC differentiation stage and continues to function as an important differentiation factor at each differentiation stage (17). Previous studies have either profiled miRNAs specifically expressed in CB cells, or investigated the role of each miRNA in the genesis of HSCs, but thus far studies concerning miRNA-induced chemotaxis of HSCs appear to be lacking (18, 19).
CXCR4 is an alpha-chemokine receptor specific for SDF-1. It is a molecule with potent chemotactic activity for lymphocytes. CXCR4 expression is either low or nonexistent in many healthy tissues, but is expressed in various cancer cells as well as in HSCs. Therefore, many studies are focused on the regulation of CXCR4 expression in order to better understand various processes associated with pathological phenomena and cell development (20).
Microarray analysis indicates that CXCR4 is a predicted target of miR-9 (21, 22). CXCR4 expression appears to be regulated by miR-9 in cancer cells (23, 24). MiR-9 has been implicated in the progression of a variety of human cancers (25, 26), where aberrant miR-9 expression is reportedly associated with many human cancers through the epigenetic inactivation of cancer-related genes (25–27). Increased CXCR4 expression in cancer cells is correlated with more distant lymphoid metastasis. Based on recent reports that miR-9 may play a role in controlling tumor trafficking and metastasis (25, 26), we hypothesized that miR-9 may regulate CXCR4-mediated chemotactic cell mobility. We tested this in CB-CD34+ cells as well as 293T and TF-1 cell lines.
We report that miR-9 may be involved in decreasing cell mobility towards SDF-1 through the down-regulation of CXCR4 expression, and that a reduction in miR-9 expression by antisense miR-9 may increase CXCR4 expression and chemotaxis. This information may be useful in efforts to improve CB-HCT.
Normal human CB was provided by CORD:USE, a cord blood banking company in Orlando, FL, and Soonchunhyang University Bucheon/Cheonan Hospital in South Korea. This study was approved by local Institutional Review Boards (IRBs; 2016-04-008-004 and 2018-05-037-002). Fully informed consent was obtained from all patients prior to donation. Mononuclear cells were isolated by density gradient centrifugation over Ficoll-Plaque Plus (GE Healthcare, Marlborough, MA, USA) according to the manufacturer’s protocol. CD34+ cells were obtained through immunomagnetic selection (Miltenyi Biotec, Auburn, CA, USA) over two sequential columns. This yielded CD34+ cells with 90–98% purity. CD34+ cells were then cultured in RPMI-1640 medium containing 10% fetal bovine serum (FBS), 100 ng/mL stem cell factor (SCF), thrombopoietin (TPO), and FMS-LIKE TYROSINE KINASE 3 LIGAND (FLT3L) to expand cell numbers (28).
Total RNA was extracted from cultured cells, using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. For the preparation of cellular miRNAs, small-sized RNAs containing miRNAs were isolated from total RNA, using the RNeasy MinElute Cleanup kit (Qiagen, Valencia, CA, USA). To evaluate miRNA expression levels, we used single-tube TaqMan miRNA assays as previously described (Applied Biosytems, Franklin Lakes, NJ, USA) (29, 30). Normalization was performed with 18S. All reverse transcription reactions, no-template controls, and RT primer minus controls were run in a GeneAmp PCR 9700 Thermocycler (Applied Biosystems). Gene expression levels were quantified using the ABI 7300 Real-Time PCR System (Applied Biosytems). Comparative real-time PCR was performed in triplicate, including the no-template controls, using specific primers for miR-9, with all reagents obtained from Applied Biosytems. The TaqMan RNU6B endogenous control kit was purchased from Applied Biosystems. PCR was carried out at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Relative expression was calculated using the comparative threshold (Ct) method.
CB-CD34+ cells were harvested at the indicated time and lysed with lysis buffer (20 mM Tris-HCl, pH 7.5, 1 mM EGTA, 1 mM EDTA, 1 mM beta-glycerol phosphate, and 1% Triton X-100; Sigma-Aldrich. St. Louis, MO, USA) containing a protease inhibitor cocktail (Roche, Ltd. Bael, Switzerland). Extracted proteins were denatured using sodium dodecyl sulfate (SDS) sample buffer at 95°C for 5 min. Proteins in the cell lysates were resolved by SDS-polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes. Immunoblotting was done using mouse anti-CXCR4 (1:1000 dilution; Santa Cruz Biotechnology, Dallas, TX, USA) and mouse anti-β-ACTIN (1:2000, Sigma-Aldrich) as primary antibodies.
The miR-9 target containing a sequence complementary to the seed sequence was selected by the miRanda (http://cbio.mskcc.org/mirnaviewer) and TargetScan (http://www.targetscan.org/) prediction programs. Wild- type and mutant 3′UTR segments of CXCR4 mRNAs were cloned into the psiCHECK2 vector containing Renilla or firefly luciferase genes as a reporter (Promega, Madison, WI, USA). The sequence, ACCAAAG, of the mutant CXCR4 3′UTR (402–408) was changed to ACCTTTC. The 293T cells were transfected with both luciferase reporter plasmids (1 μg) and 100 nM miRNA oligomers, in 3 independent replicates, and luciferase activity in the cells were assayed. Each reporter assay was conducted in triplicate.
To introduce either miR-9 or anti-miR-9 into the target cell, a commercially available lentiviral construct was purchased from SBI (PMIRH9-1PA-1 and MZIP9-PA-1). Each viral particle was obtained and transduced into each target cell following the manufacturer’s protocol. Following the isolation of CB-CD34+ cells from human cord blood, CD34+ cells were incubated with cytokine for 2 days. For western blot analysis, the cells in our CB-CD34+, TF-1, and 293 T cell lines were treated with lentiviral particles and cultured for 3 days before harvesting. Two days after transduction with lentiviral particles, the cell lines were loaded onto the upper chamber of the Transwell device and incubated for one more day in order to conduct migrating assays.
Transwell migration assays were performed as previously described, using 12-mm diameter cell culture inserts with a 5-μm pore size (Corning, Corning, NY, USA) and 12-well cell culture plates. TF-1 growth factor-dependent cells and CB-CD34+ cells (105) were transduced with miR-9, antisense miR-9 and pretreated with 10 μM AMD3100, the CXCR4 antagonist, and loaded onto the upper chamber of the Transwell device. Control medium or 100 ng/ml SDF-1 was added to the lower chamber. The cells were allowed to migrate for 24 hours in a humidified CO2 incubator at 37°C. Following incubation, the medium was aspirated and the cells that had migrated to the lower chamber were obtained and enumerated using a hematocytometer. Percentage cell migration was calculated by dividing the number of cells in the lower chamber by the total number of cells (105) and multiplying by 100.
CB-CD34+ cells contain primitive HSCs with ability for self-renewal, proliferation, and differentiation into hematopoietic cell lineages (31, 32). CB-CD34+ cells are often used for HSC transplantation. In order to increase the small number of available CB-CD34+ cells as well as to increase the proportion of HSCs in our CB-CD34+ population, we expanded the numbers of CD34+ CB cells ex-vivo with growth factors. Western blotting was performed to monitor the protein expression of CXCR4 in the CB-CD34+ cell populations expanded by the use of growth factors (100 ng/mL SCF, TPO, and FLT3L). Expression of CXCR4 protein was relatively low during early culture but started to dramatically increase after 12 hours (Fig. 1a). A noticeable increase in CXCR4 expression was seen in the 62 kDa species at 12 hours, which progressively increased to a maximum level after four days. The 62 kDa species corresponded to the ubiquitinated form of CXCR4 (33). Ubiquitin is also a natural ligand of CXCR4 (34). Ubiquitin is a small (76 amino acid) protein that is best known for its intracellular role in targeting ubiquitylated proteins for degradation via the ubiquitin proteasome system. The protein is potentially produced as a secondary cell signal by the modification of intracellular proteins. Further, the 45 kDa CXCR4 species represents a monomeric glycosylated form that is expressed similar to the ubiquitylated protein (35).
To detect miRNAs that may regulate CXCR4 expression, TargetScan analysis was performed. The analysis revealed miR-9 sequences that pair with the 3′UTR region of CXCR4 (21). CXCR4 has been identified as a target gene for miR-9 in several other computational databases and in microarray data (24). As miR-9 was highly expressed in fresh CB-CD34+ cells, we monitored miR-9 expression levels in CB-CD34+ cells using real-time PCR during the same periods of culture. The miR-9 expression pattern was inversely correlated with that of CXCR4 protein expression (Fig. 1b). Thus, in contrast to CXCR4 levels, the highest levels of miR-9 was observed in fresh CB-CD34+ cells, where these high levels gradually declined for up to four days. The inverse correlation between CXCR4 and miR-9 expression suggests that miR-9 may negatively control CXCR4 expression during HSC maturation.
Since the expressions of CXCR4 and miR-9 were mutually exclusive, we next examined whether the expression of CXCR4 was regulated by miR-9 first by using 293T cells. To verify whether miR-9 suppresses CXCR4 expression, 293T cells were co-transfected with psi-CHECK2 vector, together with either sense or antisense miR-9, following which luciferase activities were assessed. The dual-luciferase reporter vector psi-CHECK2 was fused to either wild-type 3′UTR (WT) or mutated 3′UTR (Mut) sequences. WT and Mut (ACCAAAG changed to UGGUUUC) targeted CXCR4 3′UTR sequences are depicted (Fig. 2a). Sense miR-9 significantly reduced luciferase activity when co-transfected with luciferase reporter gene fused to CXCR4 3′UTR sequences (WT). Such a miR-9 effect was not apparent when co-transfection was performed with luciferase reporter gene fused to Mut CXCR4 3′UTR (Fig. 2b). By contrast, antisense miR-9 (anti-miR-9) co-transfected with the luciferase reporter gene displayed the opposite effect on sense miR-9. Luciferase activity containing the CXCR4 3′UTR sequence (WT) was increased by anti-miR-9, but that containing mutant sequence (Mut) was not increased. These results indicate that CXCR4 3′UTR is a specific target for miR-9, and suggest that CXCR4 expression is negatively influenced by miR-9 in 293T cells.
As the CXCR4 3′UTR in the psi-CHECK2 vector was a specific target for miR-9, as noted above, through the use of luciferase activity as a reporter gene, we tested whether miR-9 can actually regulate intrinsic cellular CXCR4 protein production in primary cells as well as in other cells. To this end, we transduced CB-CD34+ cells as well as human embryonic kidney cells, 293T cells, and a growth factor-dependent human erythroleukemic cell line, TF-1. Transfection involved lentiviral vectors containing either miR-9 or antisense-miR-9. We assessed CXCR4 expression through fluorescence microscopy and western blotting, and first verified miR-9 expression through transduction with miR-9 or antisense-miR-9 by quantitative RT-PCR, where miR-9 expression in CB-CD34+ cells was significantly increased or decreased, respectively, upon their transduction (Fig. 3a). For all cells, transduction with miR-9 produced significantly lower levels of CXCR4 protein, compared to that in mock-transduced control cells (Fig. 3b–d). Notably, cells transduced with anti-miR-9 displayed a higher level of CXCR4 protein expression than control cells. Thus, miR-9 regulated expression of CXCR4 in primary CB-CD34+ cells, as well as in 293T and TF-1 cell lines, via the inhibitory and enhancing effects of miR-9 and anti-miR-9 respectively.
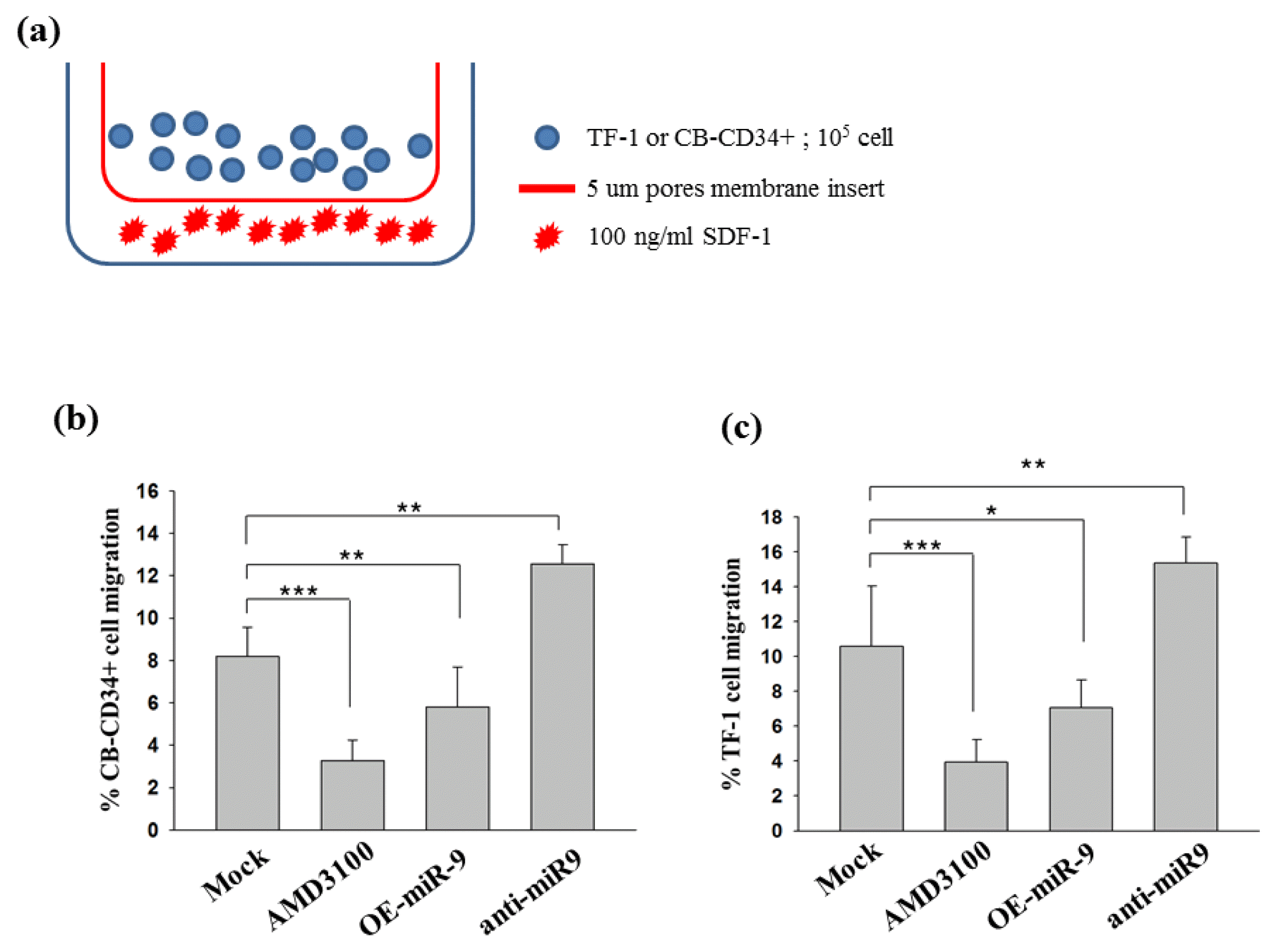
As miR-9 down-regulated CXCR4 expression, we investigated whether such down-regulation of CXCR4 expression by miR-9 corresponded to a functional outcome in the form of chemotactic cell mobility toward SDF-1, using CB-CD34+ and TF-1 cells (Fig. 4). The cells were transduced with viral vector containing either miR-9 or anti-miR-9 and placed in the upper chambers of Transwell devices (105 cells) (Fig. 4a). After 24 hours of incubation, those cells on the upper surface of the inserted membrane were gently removed using cotton tips. Cells collected from the lower chambers containing either SDF-1 (100 ng/mL) were counted using a hematocytometer, Ectopic overexpression of miR-9 in CB-CD34+ and TF-1 cells decreased chemotactic cell mobility toward SDF-1 by 29% and 35%, respectively, compared to that in mock-transduced controls (Figs. 4b and c). In contrast, anti-miR-9-transduced TF-1 and CB-CD34+ cells showed increased chemotactic activities (45% and 54%, respectively) compared to control cells. AMD3100 specifically inhibits CXCR4/SDF1 interaction in a manner which is sufficient to prevent cellular migration, indicating that miR-9 may have an effect on the regulation of cellular migration mediated via the CXCR4 receptor.
CXCR4, which is the main chemokine receptor for SDF-1, plays an important role by directing the migration of HSCs and HPCs to the BM. As CXCR4 is a potential target for miR-9 (23, 24), our study focused on monitoring the expression levels of CXCR4, in primary CB CD34+ cells, as well as several other cell lines, and the response of these cells to SDF-1-mediated chemotaxis. CXCR4 expression was relatively low in fresh CB-CD34+ cells but increased substantially as the cells were cultivated in the presence of the growth factors. CXCR4 expression levels in the cultivated CB-CD34+ cells were inversely correlated with miR-9 levels, as indicated by increased CXCR4 expression levels corresponding to decreased miR-9 expression levels. Transduction of CB-CD34+ cells and hematopoietic cancer cell lines with viral vectors to express miR-9 and anti-miR-9 demonstrated that CXCR4 expression was tightly regulated by miR-9. This observation was substantiated by the fact that ectopic expression of miR-9 inhibited cell mobility toward SDF-1 while ectopic expression of anti-miR-9 promoted cell mobility toward SDF-1. Collectively, the data suggest that miR-9 may regulate CB CD34+ cell mobility toward SDF-1. To our knowledge, this is the first report describing the role of miR-9 in hematopoietic cell mobility.
Our observations that CXCR4 expression increased in CB-CD34+ cells during cultivation is consistent with previous studies suggesting that short-term culture of CB-CD34+ cells may increase efficiency of homing to BM via CXCR4 (36). In another study, while freshly isolated CD34+CD38+ cells did not home, in vivo stimulation with granulocyte colony-stimulating factor (G-CSF) and SCF potentiated the homing abilities of cytokine-stimulated CD34+CD38+ cells (37). Additionally, grafted CB cells appear to be more CXCR4-positive than initial CD34+ cells following transplantation (3). Our findings indicate that stimulation with cytokines attenuates miR-9 production in CD34+ cells and that the reduced miR-9 expression allows for increased CXCR4 expression, and perhaps enhanced homing to BM in response to a gradient of SDF-1.
We reiterated the experiments with 293T and TF-1 cells to explore whether the effect of miR-9 on CXCR4 expression was limited to CB-CD34+ cells only, because if this effect extended to other cell lines too, the effect could be interpreted as a broader effect with universal implications. The TF-1 cell line was used to study the effect of miR-9 on cell migration, especially, since TF-1 cell lines offer a convenient system with which proliferation and differentiation of myeloid progenitor cells can be investigated. Transfection studies indicated that ectopic expression of miR-9 and anti-miR-9, down- and up- regulated CXCR4 expression, respectively (Fig. 3). Consistent with the role of miR-9 in down-regulating CXCR4 protein levels, Transwell assays with CB-CD34+ and TF-1 cells transduced with vector containing either miR-9 or anti-miR-9, indicated that miR-9 was able to inhibit cell migration toward SDF-1 (Fig. 4). In contrast, anti-miR-9 promoted migratory activities. These data suggest that miR-9 may regulate cell mobility toward SDF-1 in several types of cells. To our knowledge, ours is the first report describing the role of miR-9 in hematopoietic cell mobility.
MiR-9 is increasingly recognized as a critical regulator of organ development and neurogenesis. Reduced expression of miR-9 is considered a marker of poor survival in numerous cancers. Silencing of miR-9 by CpG island hypermethylation has been found in many tumors, clearly indicating that miR-9 possesses tumor-suppressive activities (38). However, the molecular mechanisms through which miR-9 regulates malignant phenotypes in immature hematopoietic cells are unclear. In this study, we demonstrated that miR-9 functions as a suppressor of CXCR4 expression in hematopoietic cells. Cancer stem cells also express CXCR4 on their surface, and SDF-1 is known to protect breast cancer cells from apoptosis. These facts indicate that the SDF-1/CXCR4 axis is involved in directing trafficking/metastasis in organs that highly express SDF-1. Hence, both the metastasis of cancer stem cells and trafficking of HSCs and HPCs appear to be based on similar mechanisms involving CXCR4 and miR-9.
The applicability of CB cells for HCT has been hindered by the limited number of stem cells in this population available from a single unit and their chemotactic homing capabilities (9, 10, 39). One option to enhance the use of these limited numbers of cells is to enhance CXCR4 expression in HSCs. While this option has been explored, more information on the mechanism involved in the regulation of CXCR4 expression is needed. Our data demonstrate that miR-9 is important for the regulation of CXCR4 expression in CB CD34+ cells, as well as for their chemotactic response to SDF-1. These findings, together with available information regarding the role of miR-9 in cancer cells, provide support for the postulation that up-modulation of CXCR4 expression in CD34+ cells may improve engraftment while its down-modulation in cancer cells may reduce metastasis.
These studies were supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI14C2093) as well as by the National Research Foundation of Korea (NRF-2015R1A6A1A03032522, NRF-2016R1D1A1B03931605, NRF-2018M3A9B5024060, and NRF-2017M3A9C6033069) and Global Research Development Center (NRF-2016K1A4A3914725) funded by the Ministry of Education and were supported by the Functional Districts of the Science Belt support program, Ministry of Science, ICT and Future Planning (Grant Number: 2015K000278) to MRL. Special thanks to STE-MOPIA. These studies were also supported by U.S. Public Health Service Grants from the National Institutes of Health to HEB: R01 HL056416, HL112669, R35 HL139599 and U54 DK106846.
References
1. Broxmeyer HE. Chemokines in hematopoiesis. Curr Opin Hematol. 2008; 15:49–58. DOI: 10.1097/MOH.0b013e3282f29012.

2. Kollet O, Petit I, Kahn J, Samira S, Dar A, Peled A, Deutsch V, Gunetti M, Piacibello W, Nagler A, Lapidot T. Human CD34(+)CXCR4(-) sorted cells harbor intracellular CXCR4, which can be functionally expressed and provide NOD/SCID repopulation. Blood. 2002; 100:2778–2786. DOI: 10.1182/blood-2002-02-0564. PMID: 12351385.

3. Ramirez P, Wagner JE, Defor TE, Eide CR, Miller JS, Weisdorf DJ, Brunstein CG. CXCR4 expression in CD34+ cells and unit predominance after double umbilical cord blood transplantation. Leukemia. 2013; 27:1181–1183. DOI: 10.1038/leu.2012.261. PMCID: 3668782.

4. Dar A, Goichberg P, Shinder V, Kalinkovich A, Kollet O, Netzer N, Margalit R, Zsak M, Nagler A, Hardan I, Resnick I, Rot A, Lapidot T. Chemokine receptor CXCR4-dependent internalization and resecretion of functional chemokine SDF-1 by bone marrow endothelial and stromal cells. Nat Immunol. 2006; 6:1038–1046. DOI: 10.1038/ni1251.

5. Lapidot T, Dar A, Kollet O. How do stem cells find their way home? Blood. 2005; 106:1901–1910. DOI: 10.1182/blood-2005-04-1417. PMID: 15890683.

6. Ma Q, Jones D, Borghesani PR, Segal RA, Nagasawa T, Kishimoto T, Bronson RT, Springer TA. Impaired B-lymphopoiesis, myelopoiesis, and derailed cerebellar neuron migration in CXCR4- and SDF-1-deficient mice. Proc Natl Acad Sci U S A. 1998; 95:9448–9453. DOI: 10.1073/pnas.95.16.9448. PMID: 9689100. PMCID: 21358.

7. Ponomaryov T, Peled A, Petit I, Taichman RS, Habler L, Sandbank J, Arenzana-Seisdedos F, Magerus A, Caruz A, Fujii N, Nagler A, Lahav M, Szyper-Kravitz M, Zipori D, Lapidot T. Induction of the chemokine stromal-derived factor-1 following DNA damage improves human stem cell function. J Clin Invest. 2000; 106:1331–1339. DOI: 10.1172/JCI10329. PMID: 11104786. PMCID: 381461.

8. Wysoczynski M, Reca R, Ratajczak J, Kucia M, Shirvaikar N, Honczarenko M, Mills M, Wanzeck J, Janowska-Wieczorek A, Ratajczak MZ. Incorporation of CXCR4 into membrane lipid rafts primes homing-related responses of hematopoietic stem/progenitor cells to an SDF-1 gradient. Blood. 2005; 105:40–48. DOI: 10.1182/blood-2004-04-1430.

9. Broxmeyer H. Enhancing the efficacy of engraftment of cord blood for hematopoietic cell transplantation. Transfus Apher Sci. 2016; 54:364–372. DOI: 10.1016/j.transci.2016.05.013. PMID: 27211041. PMCID: 4899263.

10. Broxmeyer HE, Farag SS, Rocha V. Cord Blood Hematopoietic Cell Transplantation. Forman SJ, Negrin RS, Antin JH, Appelbaum FR, editors. Thomas’ hematopoietic cell transplantation. Oxford: John Wiley & Sons;2016. p. 437–455. DOI: 10.1002/9781118416426.ch39.

11. Guo B, Huang X, Cooper S, Broxmeyer HE. Glucocorticoid hormone-induced chromatin remodeling enhances human hematopoietic stem cell homing and engraftment. Nat Med. 2017; 23:424–428. DOI: 10.1038/nm.4298. PMID: 28263313. PMCID: 5408457.

12. Huang X, Guo B, Liu S, Wan J, Broxmeyer HE. Neutralizing negative epigenetic regulation by HDAC5 enhances human haematopoietic stem cell homing and engraftment. Nat Commun. 2018; 9:2741. DOI: 10.1038/s41467-018-05178-5. PMID: 30013077. PMCID: 6048146.

13. Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008; 455:64–71. DOI: 10.1038/nature07242. PMID: 18668037. PMCID: 2745094.

14. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116:281–297. DOI: 10.1016/S0092-8674(04)00045-5. PMID: 14744438.
15. Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foà R, Schliwka J, Fuchs U, Novosel A, Müller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007; 129:1401–1414. DOI: 10.1016/j.cell.2007.04.040. PMID: 17604727. PMCID: 2681231.

16. Lee MR, Kim JS, Kim KS. miR-124a is important for migratory cell fate transition during gastrulation of human embryonic stem cells. Stem cells. 2010; 28:1550–1559. DOI: 10.1002/stem.490. PMID: 20665740.

17. Houshmand M, Nakhlestani Hagh M, Soleimani M, Hamidieh AA, Abroun S, Nikougoftar Zarif M. MicroRNA microarray profiling during megakaryocyte differentiation of cord blood CD133+ hematopoietic stem cells. Cell J. 2018; 20:195–203. PMID: 29633597. PMCID: 5893291.
18. Meng X, Sun B, Xue M, Xu P, Hu F, Xiao Z. Comparative analysis of microRNA expression in human mesenchymal stem cells from umbilical cord and cord blood. Genomics. 2016; 107:124–131. DOI: 10.1016/j.ygeno.2016.02.006. PMID: 26921857.

19. Merkerova M, Vasikova A, Belickova M, Bruchova H. MicroRNA expression profiles in umbilical cord blood cell lineages. Stem Cells Dev. 2010; 19:17–26. DOI: 10.1089/scd.2009.0071.

20. Peled A, Petit I, Kollet O, Magid M, Ponomaryov T, Byk T, Nagler A, Ben-Hur H, Many A, Shultz L, Lider O, Alon R, Zipori D, Lapidot T. Dependence of human stem cell engraftment and repopulation of NOD/SCID mice on CXCR4. Science. 1999; 283:845–848. DOI: 10.1126/science.283.5403.845. PMID: 9933168.

21. Bluteau O, Langlois T, Rivera-Munoz P, Favale F, Rameau P, Meurice G, Dessen P, Solary E, Raslova H, Mercher T, Debili N, Vainchenker W. Developmental changes in human megakaryopoiesis. J Thromb Haemost. 2013; 11:1730–1741. DOI: 10.1111/jth.12326. PMID: 23782903.

22. Ferrer-Marin F, Gutti R, Liu ZJ, Sola-Visner M. MiR-9 contributes to the developmental differences in CXCR-4 expression in human megakaryocytes. J Thromb Haemost. 2014; 12:282–285. DOI: 10.1111/jth.12469. PMID: 24738139. PMCID: 3989549.

23. Lu J, Luo H, Liu X, Peng Y, Zhang B, Wang L, Xu X, Peng X, Li G, Tian W, He ML, Kung H, Li XP. miR-9 targets CXCR4 and functions as a potential tumor suppressor in nasopharyngeal carcinoma. Carcinogenesis. 2014; 35:554–563. DOI: 10.1093/carcin/bgt354.

24. Yu T, Liu K, Wu Y, Fan J, Chen J, Li C, Yang Q, Wang Z. MicroRNA-9 inhibits the proliferation of oral squamous cell carcinoma cells by suppressing expression of CXCR4 via the Wnt/β-catenin signaling pathway. Oncogene. 2014; 33:5017–5027. DOI: 10.1038/onc.2013.448.

25. Chatterjee S, Behnam Azad B, Nimmagadda S. The intricate role of CXCR4 in cancer. Adv Cancer Res. 2014; 124:31–82. DOI: 10.1016/B978-0-12-411638-2.00002-1. PMID: 25287686. PMCID: 4322894.

26. Selcuklu SD, Donoghue MT, Rehmet K, de Souza Gomes M, Fort A, Kovvuru P, Muniyappa MK, Kerin MJ, Enright AJ, Spillane C. MicroRNA-9 inhibition of cell proliferation and identification of novel miR-9 targets by transcriptome profiling in breast cancer cells. J Biol Chem. 2012; 287:29516–29528. DOI: 10.1074/jbc.M111.335943. PMID: 22761433. PMCID: 3436132.

27. Sharma S, Gurudutta G. Epigenetic regulation of hematopoietic stem cells. Int J Stem Cells. 2016; 9:36–43. DOI: 10.15283/ijsc.2016.9.1.36. PMID: 27426084. PMCID: 4961102.

28. Guo B, Huang X, Lee MR, Lee SA, Broxmeyer HE. Antagonism of PPAR-γ signaling expands human hematopoietic stem and progenitor cells by enhancing glycolysis. Nat Med. 2018; 24:360–367. DOI: 10.1038/nm.4477. PMID: 29377004. PMCID: 5875416.

29. Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005; 33:e179. DOI: 10.1093/nar/gni178. PMID: 16314309. PMCID: 1292995.

30. Lee MR, Prasain N, Chae HD, Kim YJ, Mantel C, Yoder MC, Broxmeyer HE. Epigenetic regulation of NANOG by miR-302 cluster-MBD2 completes induced pluripotent stem cell reprogramming. Stem cells. 2013; 31:666–681. DOI: 10.1002/stem.1302.

31. Barker JN, Wagner JE. Umbilical cord blood transplantation: current practice and future innovations. Crit Rev Oncol Hematol. 2003; 48:35–43. DOI: 10.1016/S1040-8428(03)00092-1. PMID: 14585482.

32. Jaroscak J, Goltry K, Smith A, Waters-Pick B, Martin PL, Driscoll TA, Howrey R, Chao N, Douville J, Burhop S, Fu P, Kurtzberg J. Augmentation of umbilical cord blood (UCB) transplantation with ex vivo-expanded UCB cells: results of a phase 1 trial using the AastromReplicell System. Blood. 2003; 101:5061–5067. DOI: 10.1182/blood-2001-12-0290. PMID: 12595310.

33. Marchese A, Raiborg C, Santini F, Keen JH, Stenmark H, Benovic JL. The E3 ubiquitin ligase AIP4 mediates ubiquitination and sorting of the G protein-coupled receptor CXCR4. Dev Cell. 2003; 5:709–722. DOI: 10.1016/S1534-5807(03)00321-6. PMID: 14602072.

34. Saini V, Marchese A, Majetschak M. CXC chemokine receptor 4 is a cell surface receptor for extracellular ubiquitin. J Biol Chem. 2010; 285:15566–15576. DOI: 10.1074/jbc.M110.103408. PMID: 20228059. PMCID: 2865327.

35. Lapham CK, Romantseva T, Petricoin E, King LR, Manischewitz J, Zaitseva MB, Golding H. CXCR4 heterogeneity in primary cells: possible role of ubiquitination. J Leukoc Biol. 2002; 72:1206–1214. PMID: 12488503.
36. Ohno N, Kajiume T, Sera Y, Sato T, Kobayashi M. Short-term culture of umbilical cord blood-derived CD34 cells enhances engraftment into NOD/SCID mice through increased CXCR4 expression. Stem Cells Dev. 2009; 18:1221–1226. DOI: 10.1089/scd.2008.0298.

37. Kollet O, Spiegel A, Peled A, Petit I, Byk T, Hershkoviz R, Guetta E, Barkai G, Nagler A, Lapidot T. Rapid and efficient homing of human CD34(+)CD38(-/low)CXCR4(+) stem and progenitor cells to the bone marrow and spleen of NOD/SCID and NOD/SCID/B2m(null) mice. Blood. 2001; 97:3283–3291. DOI: 10.1182/blood.V97.10.3283. PMID: 11342460.

38. He L, Zhang L, Wang M, Wang W. miR-9 functions as a tumor inhibitor of cell proliferation in epithelial ovarian cancer through targeting the SDF-1/CXCR4 pathway. Exp Ther Med. 2017; 13:1203–1208. DOI: 10.3892/etm.2017.4118. PMID: 28413458. PMCID: 5377313.

39. Brunstein CG, Barker JN, Weisdorf DJ, DeFor TE, Miller JS, Blazar BR, McGlave PB, Wagner JE. Umbilical cord blood transplantation after nonmyeloablative conditioning: impact on transplantation outcomes in 110 adults with hematologic disease. Blood. 2007; 110:3064–3070. DOI: 10.1182/blood-2007-04-067215. PMID: 17569820. PMCID: 2018678.

Fig. 1
Increase in CXCR4 expression is related to decrease in miR-9 expression in CB-CD34+ cultures. (a) CXCR4 and actin expression levels in uncultured CB-CD34+ (lane 1), from 6 hours to 4 days (lanes 2 to 5) were assessed by western blotting (WB). Actin was used as a loading control. (b) miR-9 expression levels at defined times in CB-CD34+ cultures for 4 days as analyzed by real-time PCR. The relative expression value of miR-9 was normalized to the level of U6 mRNA. Values represent mean±SE; Student’s t-test; *p<0.05 and **p<0.01. All p-values represent the significance of miR-9 expression between 0 hour and the indicated times.
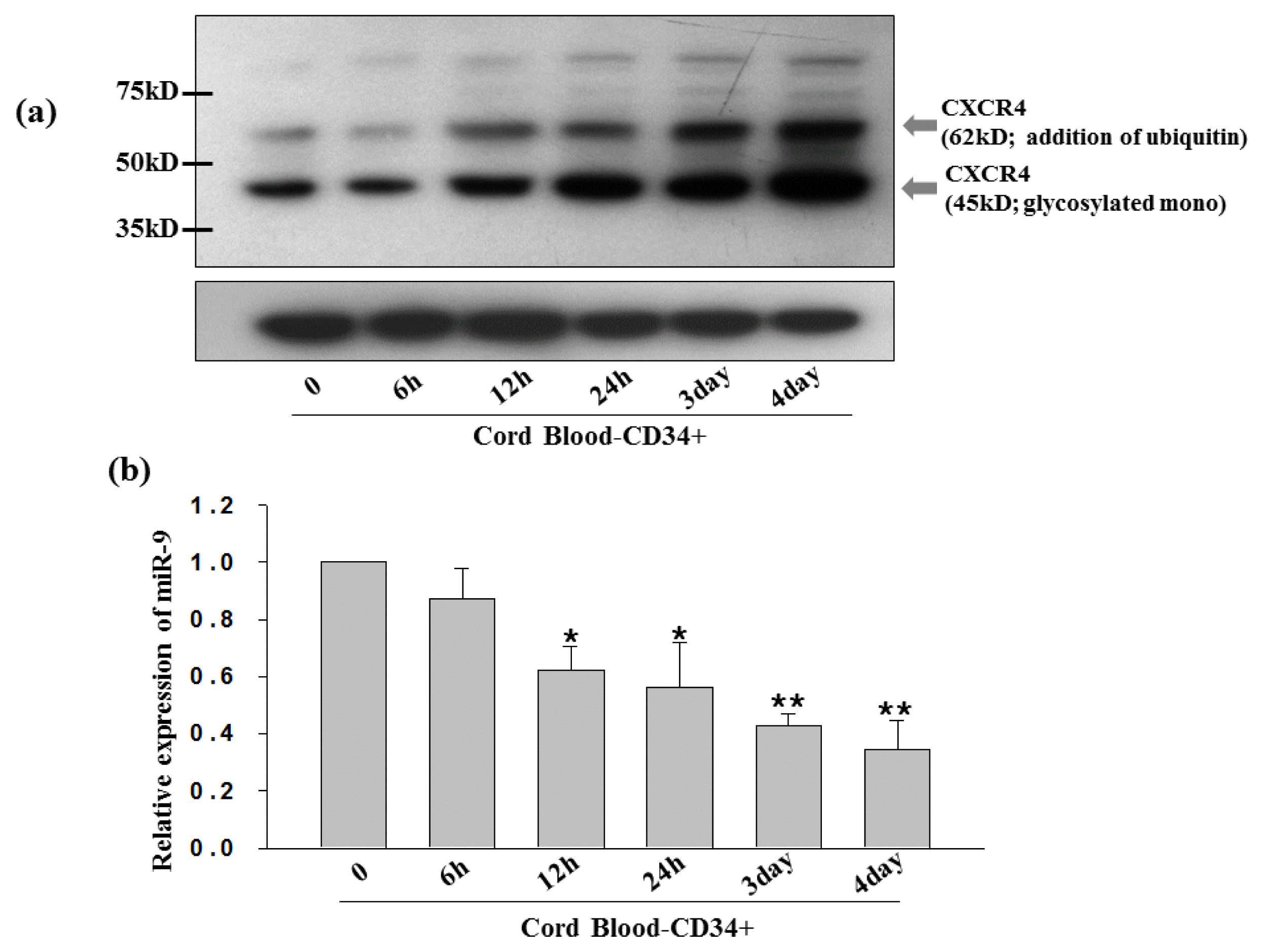
Fig. 2
miR-9 regulation of post-transcriptional CXCR4 levels. (a) Top lines depict the sequence of miR-9 predicted to have a binding site within the CXCR4 3′UTR, middle lines depict the sequence of miR-9, and bottom lines depict the sequence of the mutant CXCR4 3′ UTR. (b) 293T cells were transfected with either luciferase-reporter constructs alone (psi-CHECK) or luciferase-reporter constructs containing WT or mutant 3′UTR of CXCR4 in the presence or absence of miR-9. The effect of miR-9 on relative luciferase activities is presented as a histogram. Values represent mean±SE. Student’s t-test: **p<0.01 and ***p<0.001. All p-values represent the significance between psi-Check of luciferase activity and the treatments.
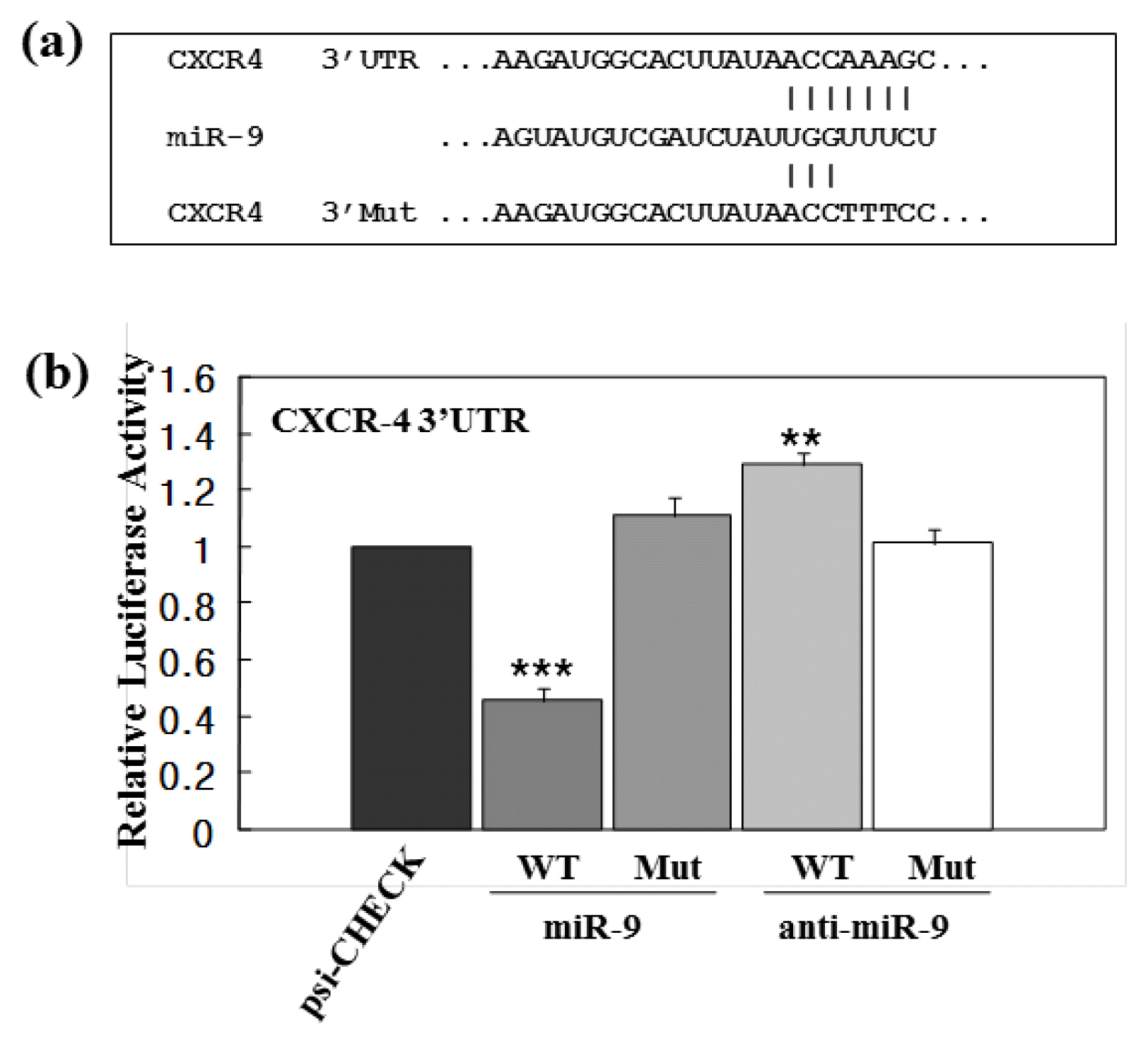
Fig. 3
miR-9 regulates CXCR4 expression. (a) CB-CD34+ cells were transduced with either miR-9 or anti-miR-9, as well as with control construct. Relative miR-9 expression levels measured by real-time PCR are displayed as histograms. Relative expression values of miR-9 are calculated by normalizing them to levels of U6 mRNA. Values represent mean±SEM. Student’s t-test: *p<0.05 and ***p<0.001. (b-d) The indicated cells were transduced with either miR-9 or anti-miR-9 as well as with the control construct. Green fluorescence indicates that transduction with miR-9 and anti-miR-9 was successful. Scale bar denotes 100 μm. The expression levels of CXCR4 in the cell lines were compared by western blotting.
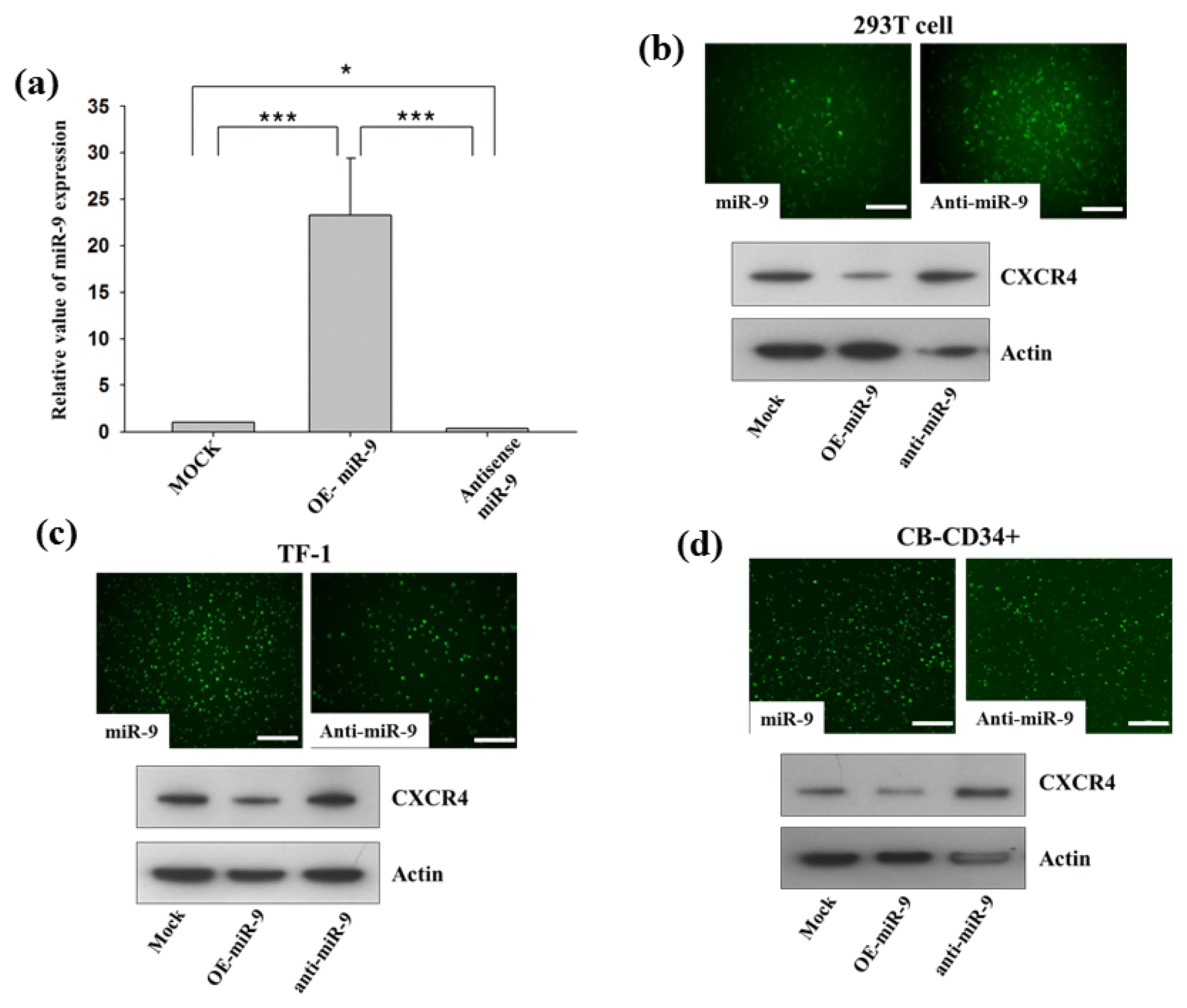
Fig. 4
miR-9 suppresses cell migration by inhibiting CXCR4. (a) The migratory cells (lower chamber) were separately harvested for the in vitro cell migration assay. In a migration assay using the Transwell system, SDF-1, which is a CXCR4 ligand, was placed in the bottom chamber of the well and 105 cells were added to the upper chamber and incubated for 24 hours. Percentage migration was calculated as (number of migrated cells/input cell number) × 100. Average cell number and standard deviation were determined from quadruplicate experiments. (b) Migration of CB-CD34+ with either miR-9 or anti-miR-9 was analyzed in the bottom chamber. Data and standard deviation were calculated from quadruplicate experiments. Student’s t-test: **p<0.01 and ***p<0.001. (c) Migration of TF-1 with either miR-9 or anti-miR-9 was analyzed in the bottom chamber. Data and standard deviation were calculated from quadruplicate experiments. Student’s t-test: *p<0.05, **p<0.01 and ***p<0.001.




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download