1. Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998; 282:1145–1147. DOI:
10.1126/science.282.5391.1145. PMID:
9804556.

2. Reubinoff BE, Pera MF, Fong CY, Trounson A, Bongso A. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat Biotechnol. 2000; 18:399–404. DOI:
10.1038/74447. PMID:
10748519.

3. Braam SR, Denning C, Matsa E, Young LE, Passier R, Mummery CL. Feeder-free culture of human embryonic stem cells in conditioned medium for efficient genetic modification. Nat Protoc. 2008; 3:1435–1443. DOI:
10.1038/nprot.2008.140. PMID:
18772870.

4. Costa M, Dottori M, Sourris K, Jamshidi P, Hatzistavrou T, Davis R, Azzola L, Jackson S, Lim SM, Pera M, Elefanty AG, Stanley EG. A method for genetic modification of human embryonic stem cells using electroporation. Nat Protoc. 2007; 2:792–796. DOI:
10.1038/nprot.2007.105. PMID:
17446878.

5. Giudice A, Trounson A. Genetic modification of human embryonic stem cells for derivation of target cells. Cell Stem Cell. 2008; 2:422–433. DOI:
10.1016/j.stem.2008.04.003. PMID:
18462693.

6. Pons J, Huang Y, Takagawa J, Arakawa-Hoyt J, Ye J, Grossman W, Kan YW, Su H. Combining angiogenic gene and stem cell therapies for myocardial infarction. J Gene Med. 2009; 11:743–753. DOI:
10.1002/jgm.1362. PMID:
19554624.

7. Rufaihah AJ, Haider HK, Heng BC, Ye L, Tan RS, Toh WS, Tian XF, Sim EK, Cao T. Therapeutic angiogenesis by transplantation of human embryonic stem cell-derived CD133+ endothelial progenitor cells for cardiac repair. Regen Med. 2010; 5:231–244. DOI:
10.2217/rme.09.83. PMID:
20210583.

8. Somers A, Jean JC, Sommer CA, Omari A, Ford CC, Mills JA, Ying L, Sommer AG, Jean JM, Smith BW, Lafyatis R, Demierre MF, Weiss DJ, French DL, Gadue P, Murphy GJ, Mostoslavsky G, Kotton DN. Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem Cells. 2010; 28:1728–1740. DOI:
10.1002/stem.495. PMID:
20715179. PMCID:
3422663.

9. Zwaka TP, Thomson JA. Homologous recombination in human embryonic stem cells. Nat Biotechnol. 2003; 21:319–321. DOI:
10.1038/nbt788. PMID:
12577066.

10. Braam SR, Denning C, van den Brink S, Kats P, Hochstenbach R, Passier R, Mummery CL. Improved genetic manipulation of human embryonic stem cells. Nat Methods. 2008; 5:389–392. DOI:
10.1038/nmeth.1200. PMID:
18391958.

11. Eiges R, Schuldiner M, Drukker M, Yanuka O, Itskovitz-Eldor J, Benvenisty N. Establishment of human embryonic stem cell-transfected clones carrying a marker for undifferentiated cells. Curr Biol. 2001; 11:514–518. DOI:
10.1016/S0960-9822(01)00144-0. PMID:
11413002.

12. Gerrard L, Zhao D, Clark AJ, Cui W. Stably transfected human embryonic stem cell clones express OCT4-specific green fluorescent protein and maintain self-renewal and pluripotency. Stem Cells. 2005; 23:124–133. DOI:
10.1634/stemcells.2004-0102.

13. Hohenstein KA, Pyle AD, Chern JY, Lock LF, Donovan PJ. Nucleofection mediates high-efficiency stable gene knockdown and transgene expression in human embryonic stem cells. Stem Cells. 2008; 26:1436–1443. DOI:
10.1634/stemcells.2007-0857. PMID:
18323409. PMCID:
3882114.

14. Yao S, Sukonnik T, Kean T, Bharadwaj RR, Pasceri P, Ellis J. Retrovirus silencing, variegation, extinction, and memory are controlled by a dynamic interplay of multiple epigenetic modifications. Mol Ther. 2004; 10:27–36. DOI:
10.1016/j.ymthe.2004.04.007. PMID:
15233939.

15. Villa-Diaz LG, Garcia-Perez JL, Krebsbach PH. Enhanced transfection efficiency of human embryonic stem cells by the incorporation of DNA liposomes in extracellular matrix. Stem Cells Dev. 2010; 19:1949–1957. DOI:
10.1089/scd.2009.0505. PMID:
20367256. PMCID:
3109506.

16. Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, Smuga-Otto K, Howden SE, Diol NR, Propson NE, Wagner R, Lee GO, Antosiewicz-Bourget J, Teng JM, Thomson JA. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011; 8:424–429. DOI:
10.1038/nmeth.1593. PMID:
21478862. PMCID:
3084903.

17. Mikkola M, Olsson C, Palgi J, Ustinov J, Palomaki T, Horelli-Kuitunen N, Knuutila S, Lundin K, Otonkoski T, Tuuri T. Distinct differentiation characteristics of individual human embryonic stem cell lines. BMC Dev Biol. 2006; 6:40. DOI:
10.1186/1471-213X-6-40. PMID:
16895598. PMCID:
1557488.
18. Spits C, Mateizel I, Geens M, Mertzanidou A, Staessen C, Vandeskelde Y, Van der Elst J, Liebaers I, Sermon K. Recurrent chromosomal abnormalities in human embryonic stem cells. Nat Biotechnol. 2008; 26:1361–1363. DOI:
10.1038/nbt.1510. PMID:
19029912.

19. Beers J, Gulbranson DR, George N, Siniscalchi LI, Jones J, Thomson JA, Chen G. Passaging and colony expansion of human pluripotent stem cells by enzyme-free dissociation in chemically defined culture conditions. Nat Protoc. 2012; 7:2029–2040. DOI:
10.1038/nprot.2012.130. PMID:
23099485. PMCID:
3571618.

20. Irion S, Luche H, Gadue P, Fehling HJ, Kennedy M, Keller G. Identification and targeting of the ROSA26 locus in human embryonic stem cells. Nat Biotechnol. 2007; 25:1477–1482. DOI:
10.1038/nbt1362. PMID:
18037879.

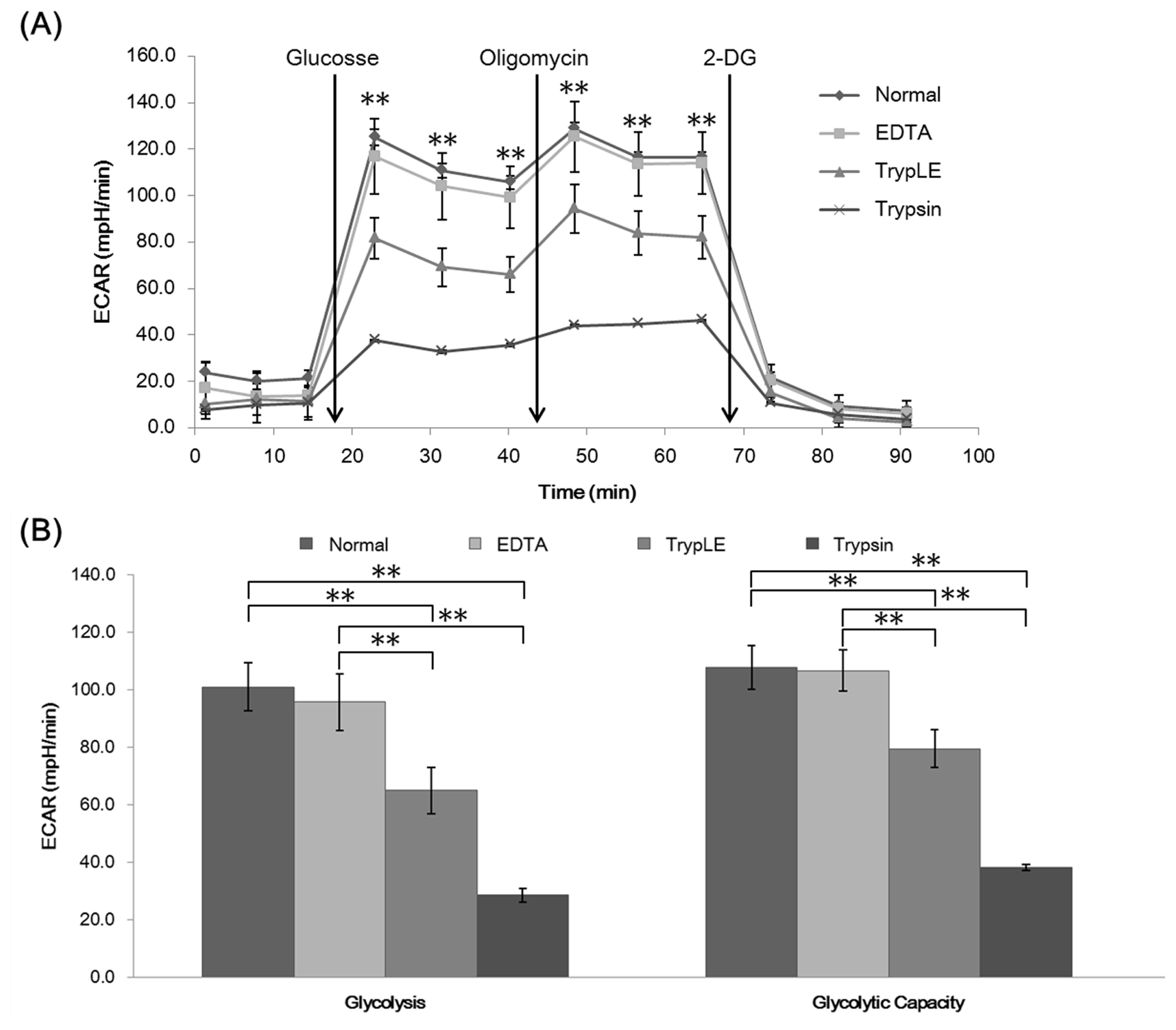
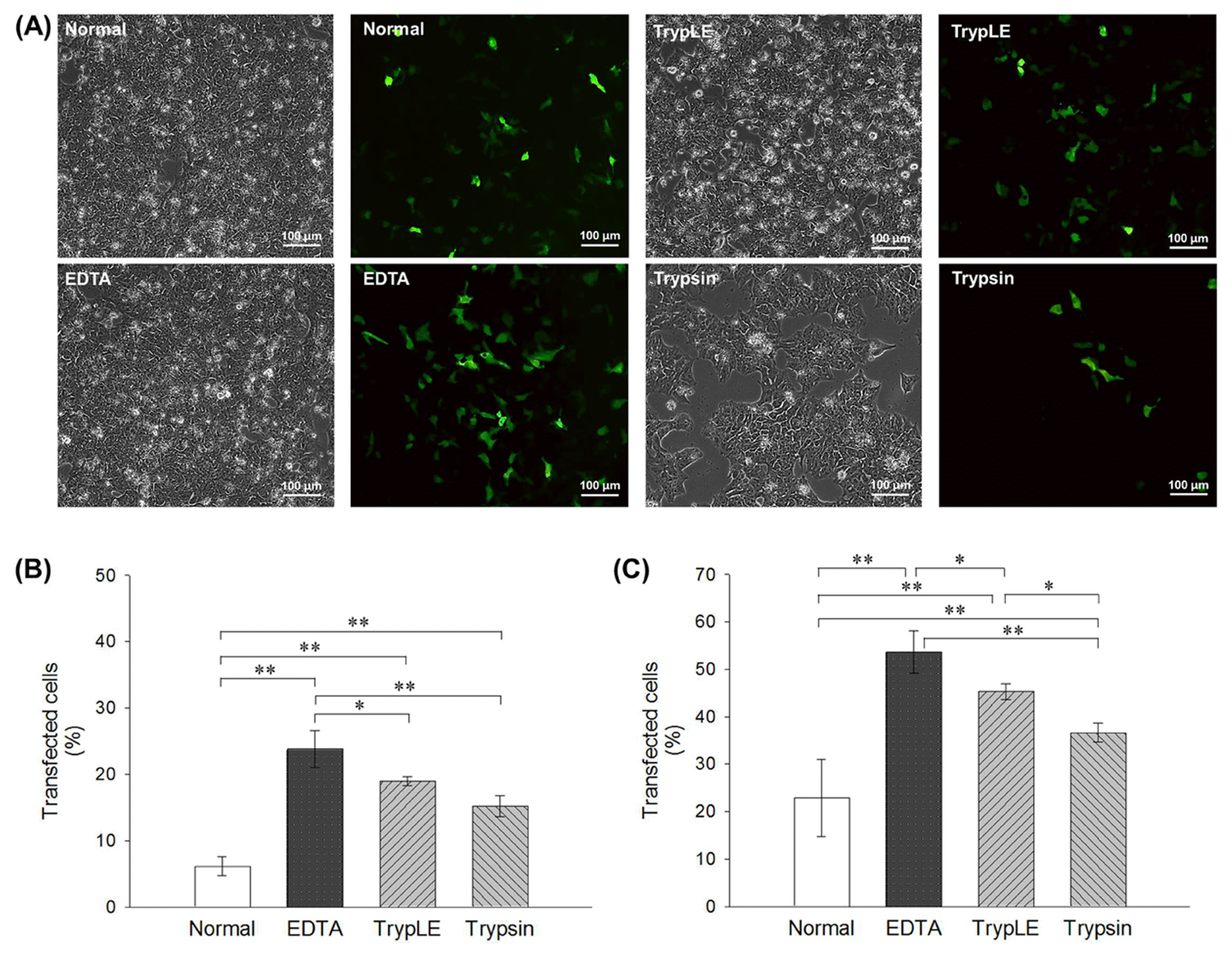
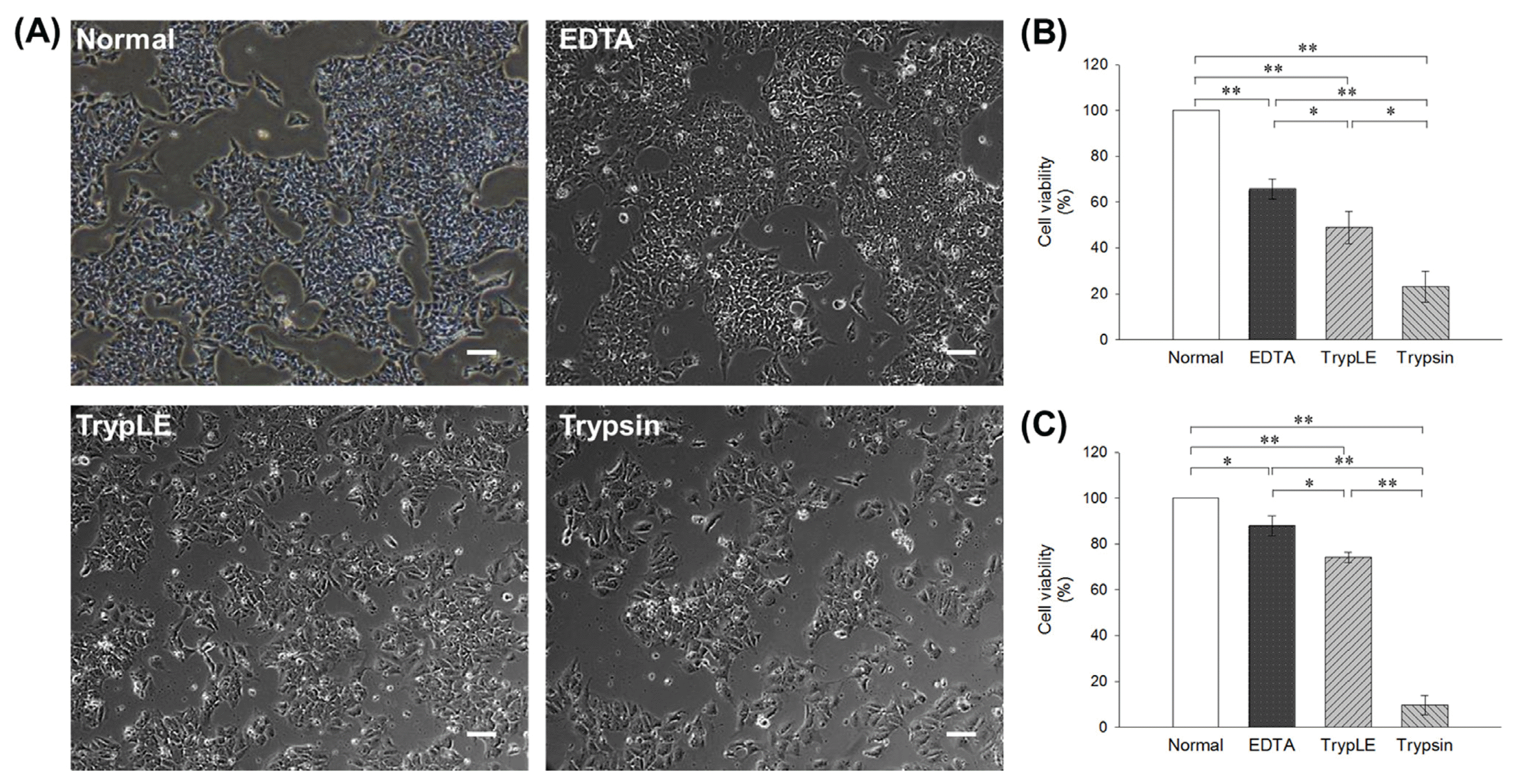
21. Badur MG, Zhang H, Metallo CM. Enzymatic passaging of human embryonic stem cells alters central carbon metabolism and glycan abundance. Biotechnol J. 2015; 10:1600–1611. DOI:
10.1002/biot.201400749. PMID:
26289220. PMCID:
4885641.





 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download