Introduction
Materials and Methods
Mouse strains and treatment
Immunofluorescence (IF) staining
EYFP+ cell isolation and organoid culture
Intestinal organoid paraffin section preparation
Statistical analysis
Results and Discussion
KLF4 is expressed in goblet cells in the crypt of small intestine
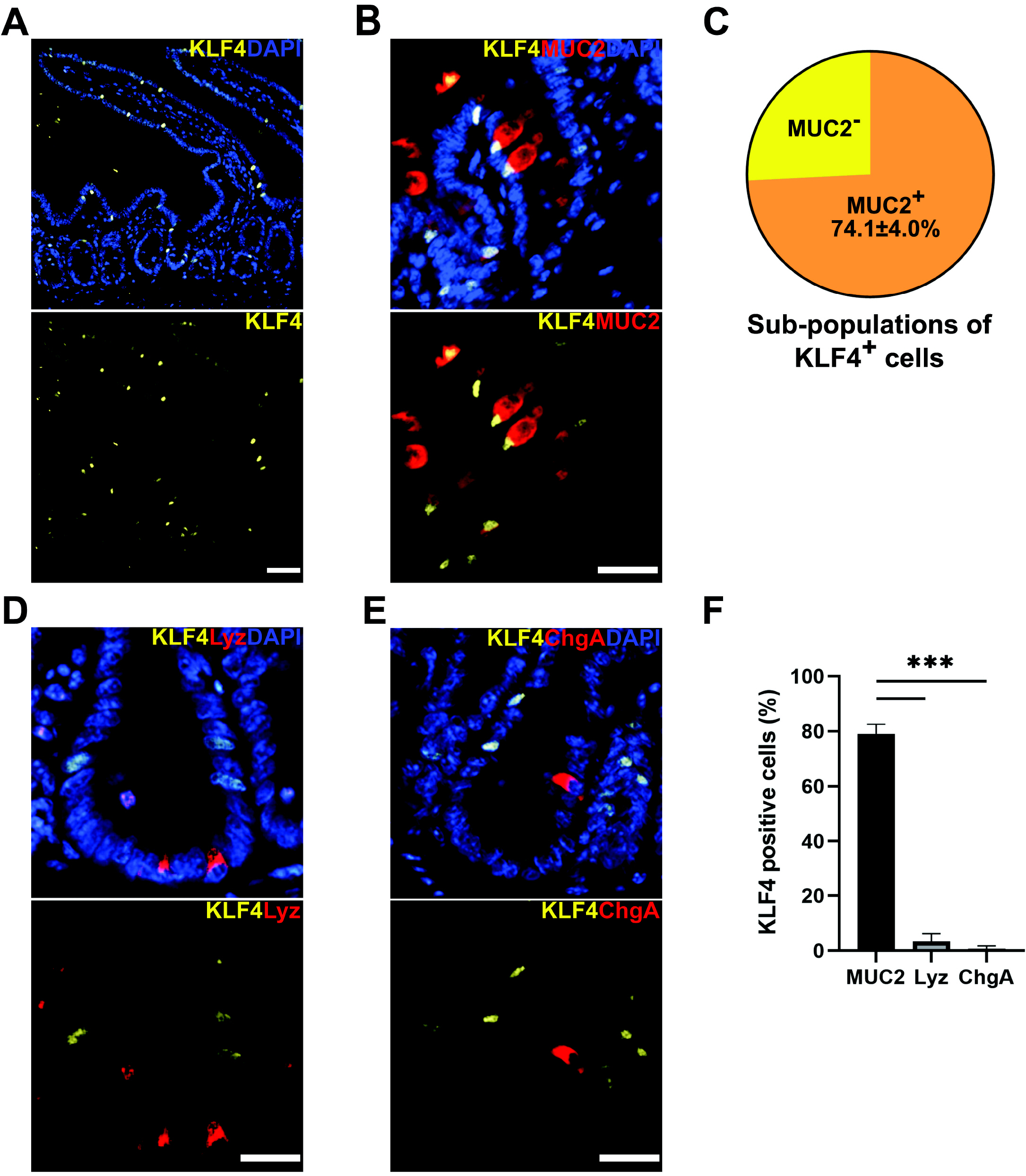 | Fig. 1Relationship between KLF4 expression and markers of differentiated cells in proximal small intestine. (A) Immunostaining of Bmi1Ctrl mouse intestine. KLF4 (yellow) was expressed mainly in the terminally differentiated epithelial cells and was also expressed in a subpopulation of the cells in the crypt. Scale bar, 50 μm. (B) Immunostaining for MUC2 (goblet cell marker, red) and KLF4 (yellow). MUC2 was overlapped with KLF4 in the crypt of proximal small intestine. Scale bar, 25 μm. (C) MUC2 was co-expressed in 74.1±4.0% of KLF4+ cells. KLF4+ cells were collected from a total of 50 crypts per Bmi1Ctrl mouse (n=3). (D) Immunostaining for lysozyme (Paneth cell marker, red) and KLF4 (yellow). Scale bar, 25 μm. (E) Immunostaining for chromogranin A (enteroendocrine cell marker, red) and KLF4 (yellow). Scale bar, 25 μm. (F) Co-expression rate of KLF4+ in MUC2+ cells was significantly higher than Lyz+ and ChgA+ cells (79.0±2.0%, 3.3±1.7%, and 0.7±0.7%, respectively). ***p<0.001 MUC2+, lysozyme+, and chromogranin A+ cells were collected from a total of 150, 100 and 60 crypts from 3 Bmi1Ctrl mice, respectively (n=3 for each group). Lyz, lysozyme; ChgA, chromogranin A. |
KLF4 regulates goblet cell differentiation and proliferation in BMI1+-ISC lineage in vivo
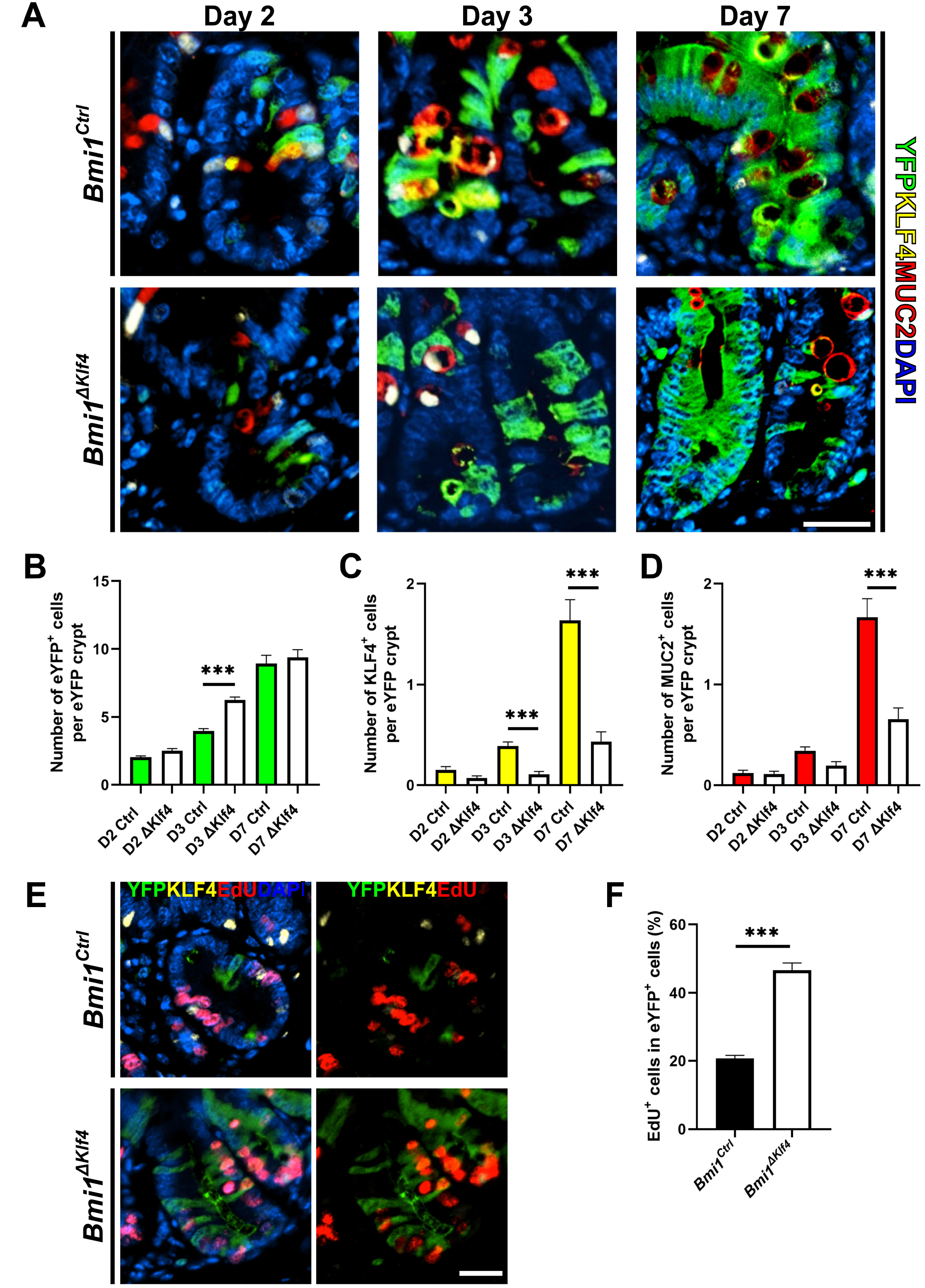 | Fig. 2KLF4 modulates goblet cell differentiation in Bmi1 cell lineage during homeostasis. (A) Immunostaining for eYFP (green), MUC2 (red) and KLF4 (yellow). Representative image of proximal small intestinal crypt at day 2, 3 and 7 following tamoxifen injection. Goblet cell differentiation is involved in Bmi1-eYFP cell lineage. Scale bar, 25 μm. (B), (C) and (D) The number of eYFP, MUC2 and KLF4 positive cells per eYFP+ including crypt increased as the time following tamoxifen administration in both groups. Data were collected from at least 20 crypts per mouse (n=3 each group). Dunn’s multiple comparisons test with Kruskal-Wallis test. (B) The number of eYFP+ cells was significantly higher in Bmi1ΔKlf4 than in Bmi1Ctrl at day 3 following tamoxifen administration. (C) The number of KLF4+ cells was significantly higher in Bmi1Ctrl than in Bmi1ΔKlf4 at day 7 following tamoxifen administration (p<0.001). (D) The number of MUC2+ cells was significantly lower in Bmi1ΔKlf4 than in Bmi1Ctrl at day 3 and 7 following tamoxifen administration (p<0.001). (E) Immunostaining for eYFP (green), KLF4 (yellow) and EdU (red) for the proximal small intestinal crypts at day 3 following tamoxifen administration. Scale bar, 25 μm. (F) At day 3 after tamoxifen injection, EdU positivity in eYFP+ cells of Bmi1ΔKlf4 was significantly higher than Bmi1Ctrl (46.7±2.1% and 20.7±0.9%, respectively). Student t test. EYFP+ cells were collected from 50 crypts per mouse (n=3 each group). ***p<0.001. |
KLF4 is positively correlated with goblet cell differentiation
 | Fig. 3KLF4 is positively correlated with goblet cell differentiation. (A) The pie chart shows the subpopula-tions of eYFP+ cells distinguished by co-staining with KLF4 and/or MUC2. A total of at 150∼260 eYFP+ cells were counted from each mouse at day 7 after tamoxifen administration (n=3 each group). (B) KLF4 and MUC2 were significantly positively correlated (Spearman correlation= 0.9429, p<0.01). |
Goblet cell differentiation from single sorted Bmi1-eYFP+ cell is regulated by KLF4
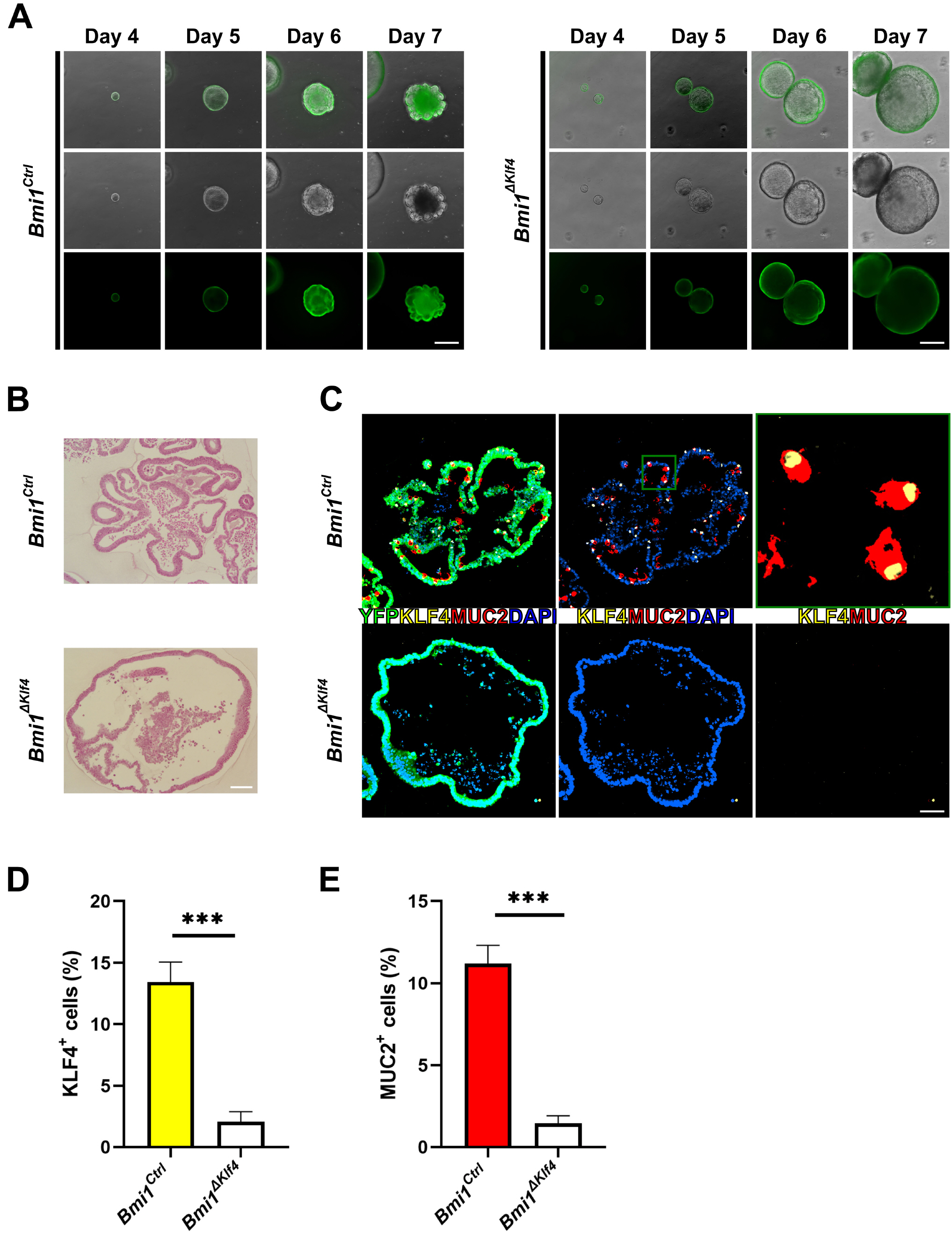 | Fig. 4Organoids derived from single FACS sorted eYFP+ cells of Bmi1Ctrl and Bmi1ΔKlf4 mice. (A) Representative time course images of cultured organoids derived from Bmi1Ctrl and Bmi1ΔKlf4 mice. Upper panels, merged image of bright-field and fluorescent image; middle panel, bright field image; lower panel, fluorescent image. Scale bars, 500 μm. (B) Hematoxylin and eosin staining at culture day 7. Scale bars, 100 μm. (C) Cultured organoids were collected at day 7. Immunostaining for eYFP (green), MUC2 (red), KLF4 (yellow). Left panel merged image for eYFP, MUC2, KLF4 and DAPI (blue); middle panel, merged image for MUC2, KLF4 and DAPI; right panel merged image for MUC2 and KLF4. Upper right panel is magnified image of green square in upper middle panel. In Bmi1Ctrl mice derived organoids, 73.9% of MUC2+ cells were co-expressed with KLF4. Scale bars, 50 μm. (D) and (E) Percentage of KLF4+ and MUC2+ cells were significantly higher in Bmi1Ctrl organoid than Bmi1ΔKlf4 at culture day 7 (Bmi1Ctrl vs. Bmi1ΔKlf4, 11.2±1.1% vs. 1.4±0.5% and 13.4±1.6% vs 2.1±0.8% for KLF4 and MUC2, respectively). Cells were analyzed from 10 sections for each group and 10∼200 cells were collected from each section. Organoids were pooled from 3 mice for each group. ***p<0.001. |




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download