1. Sakai D, Andersson GB. 2015; Stem cell therapy for intervertebral disc regeneration: obstacles and solutions. Nat Rev Rheumatol. 11:243–256. DOI:
10.1038/nrrheum.2015.13. PMID:
25708497.

2. Breivik H, Eisenberg E, O'Brien T. 2013; The individual and societal burden of chronic pain in Europe: the case for strategic prioritisation and action to improve knowledge and availability of appropriate care. BMC Public Health. 13:1229. DOI:
10.1186/1471-2458-13-1229. PMID:
24365383. PMCID:
PMC3878786.

3. Wang F, Shi R, Cai F, Wang YT, Wu XT. 2015; Stem cell approaches to intervertebral disc regeneration: obstacles from the disc microenvironment. Stem Cells Dev. 24:2479–2495. DOI:
10.1089/scd.2015.0158. PMID:
26228642.

4. Omlor GW, Fischer J, Kleinschmitt K, Benz K, Holschbach J, Brohm K, Anton M, Guehring T, Richter W. 2014; Short-term follow-up of disc cell therapy in a porcine nucleotomy model with an albumin-hyaluronan hydrogel: in vivo and in vitro results of metabolic disc cell activity and implant distribution. Eur Spine J. 23:1837–1847. DOI:
10.1007/s00586-014-3314-y. PMID:
24801573.

5. Tam V, Rogers I, Chan D, Leung VY, Cheung KM. 2014; A comparison of intravenous and intradiscal delivery of multipotential stem cells on the healing of injured intervertebral disk. J Orthop Res. 32:819–825. DOI:
10.1002/jor.22605. PMID:
24578095.

6. Wuertz K, Godburn K, Neidlinger-Wilke C, Urban J, Iatridis JC. 2008; Behavior of mesenchymal stem cells in the chemical microenvironment of the intervertebral disc. Spine (Phila Pa 1976). 33:1843–1849. DOI:
10.1097/BRS.0b013e31817b8f53. PMID:
18670337. PMCID:
PMC2567058.

7. Liang C, Li H, Tao Y, Zhou X, Li F, Chen G, Chen Q. 2012; Responses of human adipose-derived mesenchymal stem cells to chemical microenvironment of the intervertebral disc. J Transl Med. 10:49. DOI:
10.1186/1479-5876-10-49. PMID:
22424131. PMCID:
PMC3338074.

8. Johnson WE, Stephan S, Roberts S. 2008; The influence of serum, glucose and oxygen on intervertebral disc cell growth in vitro: implications for degenerative disc disease. Arthritis Res Ther. 10:R46. DOI:
10.1186/ar2405. PMID:
18433481. PMCID:
PMC2453766.

9. Erwin WM, Islam D, Eftekarpour E, Inman RD, Karim MZ, Fehlings MG. 2013; Intervertebral disc-derived stem cells: implications for regenerative medicine and neural repair. Spine (Phila Pa 1976). 38:211–216. DOI:
10.1097/BRS.0b013e318266a80d. PMID:
22772571.

10. Risbud MV, Guttapalli A, Tsai TT, Lee JY, Danielson KG, Vaccaro AR, Albert TJ, Gazit Z, Gazit D, Shapiro IM. 2007; Evidence for skeletal progenitor cells in the degenerate human intervertebral disc. Spine (Phila Pa 1976). 32:2537–2544. DOI:
10.1097/BRS.0b013e318158dea6. PMID:
17978651.

11. Mizrahi O, Sheyn D, Tawackoli W, Ben-David S, Su S, Li N, Oh A, Bae H, Gazit D, Gazit Z. 2013; Nucleus pulposus degeneration alters properties of resident progenitor cells. Spine J. 13:803–814. DOI:
10.1016/j.spinee.2013.02.065. PMID:
23578990. PMCID:
PMC3759825.

12. Han B, Wang HC, Li H, Tao YQ, Liang CZ, Li FC, Chen G, Chen QX. 2014; Nucleus pulposus mesenchymal stem cells in acidic conditions mimicking degenerative intervertebral discs give better performance than adipose tissue-derived mesenchymal stem cells. Cells Tissues Organs. 199:342–352. DOI:
10.1159/000369452. PMID:
25661884.

13. Li H, Tao Y, Liang C, Han B, Li F, Chen G, Chen Q. 2013; Influence of hypoxia in the intervertebral disc on the biological behaviors of rat adipose- and nucleus pulposus-derived mesenchymal stem cells. Cells Tissues Organs. 198:266–277. DOI:
10.1159/000356505. PMID:
24356285.

14. Tao YQ, Liang CZ, Li H, Zhang YJ, Li FC, Chen G, Chen QX. 2013; Potential of co-culture of nucleus pulposus mesenchymal stem cells and nucleus pulposus cells in hyperosmotic microenvironment for intervertebral disc regeneration. Cell Biol Int. 37:826–834. DOI:
10.1002/cbin.10110. PMID:
23554141.

15. Naqvi SM, Buckley CT. 2015; Differential response of encapsulated nucleus pulposus and bone marrow stem cells in isolation and coculture in alginate and chitosan hydrogels. Tissue Eng Part A. 21:288–299. DOI:
10.1089/ten.tea.2013.0719. PMID:
25060596.

16. Richardson SM, Hoyland JA. 2008; Stem cell regeneration of degenerated intervertebral discs: current status. Curr Pain Headache Rep. 12:83–88. DOI:
10.1007/s11916-008-0016-3. PMID:
18474185.

17. Wu J, Wang D, Ruan D, He Q, Zhang Y, Wang C, Xin H, Xu C, Liu Y. 2014; Prolonged expansion of human nucleus pulposus cells expressing human telomerase reverse transcriptase mediated by lentiviral vector. J Orthop Res. 32:159–166. DOI:
10.1002/jor.22474. PMID:
23983186.

18. Le Maitre CL, Baird P, Freemont AJ, Hoyland JA. 2009; An in vitro study investigating the survival and phenotype of mesenchymal stem cells following injection into nucleus pulposus tissue. Arthritis Res Ther. 11:R20. DOI:
10.1186/ar2611. PMID:
19210770. PMCID:
PMC2688252.

19. Saeed H, Ahsan M, Saleem Z, Iqtedar M, Islam M, Danish Z, Khan AM. 2016; Mesenchymal stem cells (MSCs) as skeletal therapeutics-an update. J Biomed Sci. 23:41. DOI:
10.1186/s12929-016-0254-3. PMID:
27084089. PMCID:
PMC4833928.

20. Chen L, Xu Y, Zhao J, Zhang Z, Yang R, Xie J, Liu X, Qi S. 2014; Conditioned medium from hypoxic bone marrow-derived mesenchymal stem cells enhances wound healing in mice. PLoS One. 9:e96161. DOI:
10.1371/journal.pone.0096161. PMID:
24781370. PMCID:
PMC4004560.

21. Hou C, Shen L, Huang Q, Mi J, Wu Y, Yang M, Zeng W, Li L, Chen W, Zhu C. 2013; The effect of heme oxygenase-1 complexed with collagen on MSC performance in the treatment of diabetic ischemic ulcer. Biomaterials. 34:112–120. DOI:
10.1016/j.biomaterials.2012.09.022. PMID:
23059006.

22. Sun Z, Liu ZH, Zhao XH, Sun L, Chen YF, Zhang WL, Gao Y, Zhang YZ, Wan ZY, Samartzis D, Wang HQ, Luo ZJ. 2013; Impact of direct cell co-cultures on human adipose-derived stromal cells and nucleus pulposus cells. J Orthop Res. 31:1804–1813. DOI:
10.1002/jor.22439. PMID:
23913869.

23. Yamamoto Y, Mochida J, Sakai D, Nakai T, Nishimura K, Kawada H, Hotta T. 2004; Upregulation of the viability of nucleus pulposus cells by bone marrow-derived stromal cells: significance of direct cell-to-cell contact in coculture system. Spine (Phila Pa 1976). 29:1508–1514. DOI:
10.1097/01.BRS.0000131416.90906.20. PMID:
15247571.

24. Sakai D, Mochida J, Yamamoto Y, Nomura T, Okuma M, Nishimura K, Nakai T, Ando K, Hotta T. 2003; Transplantation of mesenchymal stem cells embedded in Atelocollagen gel to the intervertebral disc: a potential therapeutic model for disc degeneration. Biomaterials. 24:3531–3541. DOI:
10.1016/S0142-9612(03)00222-9.

25. Wu H, Zeng X, Yu J, Shang Y, Tu M, Cheang LH, Zhang J. 2017; Comparison of nucleus pulposus stem/progenitor cells isolated from degenerated intervertebral discs with umbilical cord derived mesenchymal stem cells. Exp Cell Res. 361:324–332. DOI:
10.1016/j.yexcr.2017.10.034. PMID:
29097182.

26. Wu H, Shang Y, Zhang J, Cheang LH, Zeng X, Tu M. 2017; The effects of liquid crystal-based composite substrates on cell functional responses of human umbilical cord-derived mesenchymal stem cells by mechano-regulatory process. J Biomater Appl. 32:492–503. DOI:
10.1177/0885328217733378. PMID:
28992805.

27. Richardson SM, Kalamegam G, Pushparaj PN, Matta C, Memic A, Khademhosseini A, Mobasheri R, Poletti FL, Hoyland JA, Mobasheri A. 2016; Mesenchymal stem cells in regenerative medicine: focus on articular cartilage and intervertebral disc regeneration. Methods. 99:69–80. DOI:
10.1016/j.ymeth.2015.09.015. PMID:
26384579.

28. Katsha AM, Ohkouchi S, Xin H, Kanehira M, Sun R, Nukiwa T, Saijo Y. 2011; Paracrine factors of multipotent stromal cells ameliorate lung injury in an elastase-induced emphysema model. Mol Ther. 19:196–203. DOI:
10.1038/mt.2010.192. PMID:
20842104. PMCID:
PMC3017437.

30. Shafei AE, Ali MA, Ghanem HG, Shehata AI, Abdelgawad AA, Handal HR, Talaat KA, Ashaal AE, El-Shal AS. 2017; Mesenchymal stem cell therapy: a promising cell-based therapy for treatment of myocardial infarction. J Gene Med. 19:e2995. DOI:
10.1002/jgm.2995. PMID:
29044850.

31. Fellows CR, Matta C, Zakany R, Khan IM, Mobasheri A. 2016; Adipose, bone marrow and synovial joint-derived mesenchymal stem cells for cartilage repair. Front Genet. 7:213. DOI:
10.3389/fgene.2016.00213. PMID:
28066501. PMCID:
PMC5167763.

32. Di Rocco G, Baldari S, Toietta G. 2016; Towards therapeutic delivery of extracellular vesicles: strategies for in vivo tracking and biodistribution analysis. Stem Cells Int. 2016:5029619. DOI:
10.1155/2016/5029619. PMID:
27994623. PMCID:
PMC5141304.
33. Fong CY, Tam K, Cheyyatraivendran S, Gan SU, Gauthaman K, Armugam A, Jeyaseelan K, Choolani M, Biswas A, Bongso A. 2014; Human Wharton's jelly stem cells and its conditioned medium enhance healing of excisional and diabetic wounds. J Cell Biochem. 115:290–302. DOI:
10.1002/jcb.24661. PMID:
24038311.

34. Shen C, Lie P, Miao T, Yu M, Lu Q, Feng T, Li J, Zu T, Liu X, Li H. 2015; Conditioned medium from umbilical cord mesenchymal stem cells induces migration and angiogenesis. Mol Med Rep. 12:20–30. DOI:
10.3892/mmr.2015.3409. PMID:
25739039. PMCID:
PMC4438972.

35. Walter MN, Wright KT, Fuller HR, MacNeil S, Johnson WE. 2010; Mesenchymal stem cell-conditioned medium accelerates skin wound healing: an in vitro study of fibroblast and keratinocyte scratch assays. Exp Cell Res. 316:1271–1281. DOI:
10.1016/j.yexcr.2010.02.026. PMID:
20206158.

36. Sakai D, Nakamura Y, Nakai T, Mishima T, Kato S, Grad S, Alini M, Risbud MV, Chan D, Cheah KS, Yamamura K, Masuda K, Okano H, Ando K, Mochida J. 2012; Exhaustion of nucleus pulposus progenitor cells with ageing and degeneration of the intervertebral disc. Nat Commun. 3:1264. DOI:
10.1038/ncomms2226. PMID:
23232394. PMCID:
PMC3535337.

37. Seo KW, Lee SR, Bhandari DR, Roh KH, Park SB, So AY, Jung JW, Seo MS, Kang SK, Lee YS, Kang KS. 2009; OCT4A contributes to the stemness and multi-potency of human umbilical cord blood-derived multipotent stem cells (hUCB-MSCs). Biochem Biophys Res Commun. 384:120–125. DOI:
10.1016/j.bbrc.2009.04.094. PMID:
19394308.

38. Tsai CC, Su PF, Huang YF, Yew TL, Hung SC. 2012; Oct4 and Nanog directly regulate Dnmt1 to maintain self-renewal and undifferentiated state in mesenchymal stem cells. Mol Cell. 47:169–182. DOI:
10.1016/j.molcel.2012.06.020. PMID:
22795133.

39. Li CY, Wu XY, Tong JB, Yang XX, Zhao JL, Zheng QF, Zhao GB, Ma ZJ. 2015; Comparative analysis of human mesenchymal stem cells from bone marrow and adipose tissue under xeno-free conditions for cell therapy. Stem Cell Res Ther. 6:55. DOI:
10.1186/s13287-015-0066-5. PMID:
25884704. PMCID:
PMC4453294.

40. Song K, Gu T, Shuang F, Tang J, Ren D, Qin J, Hou S. 2015; Adipose-derived stem cells improve the viability of nucleus pulposus cells in degenerated intervertebral discs. Mol Med Rep. 12:4664–4668. DOI:
10.3892/mmr.2015.3895. PMID:
26059030.

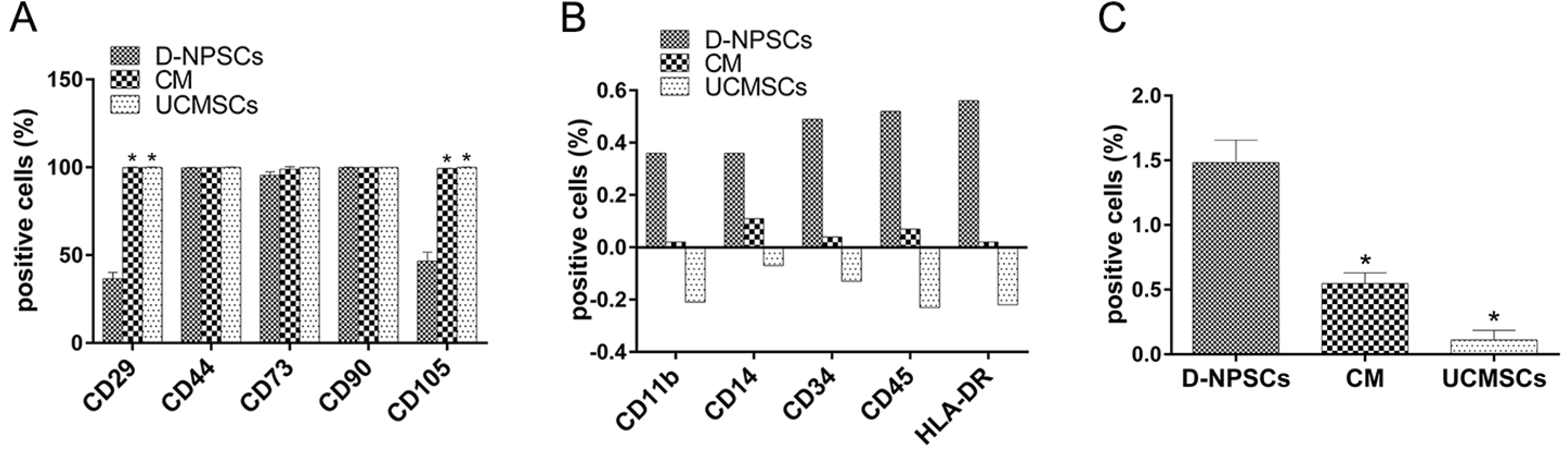
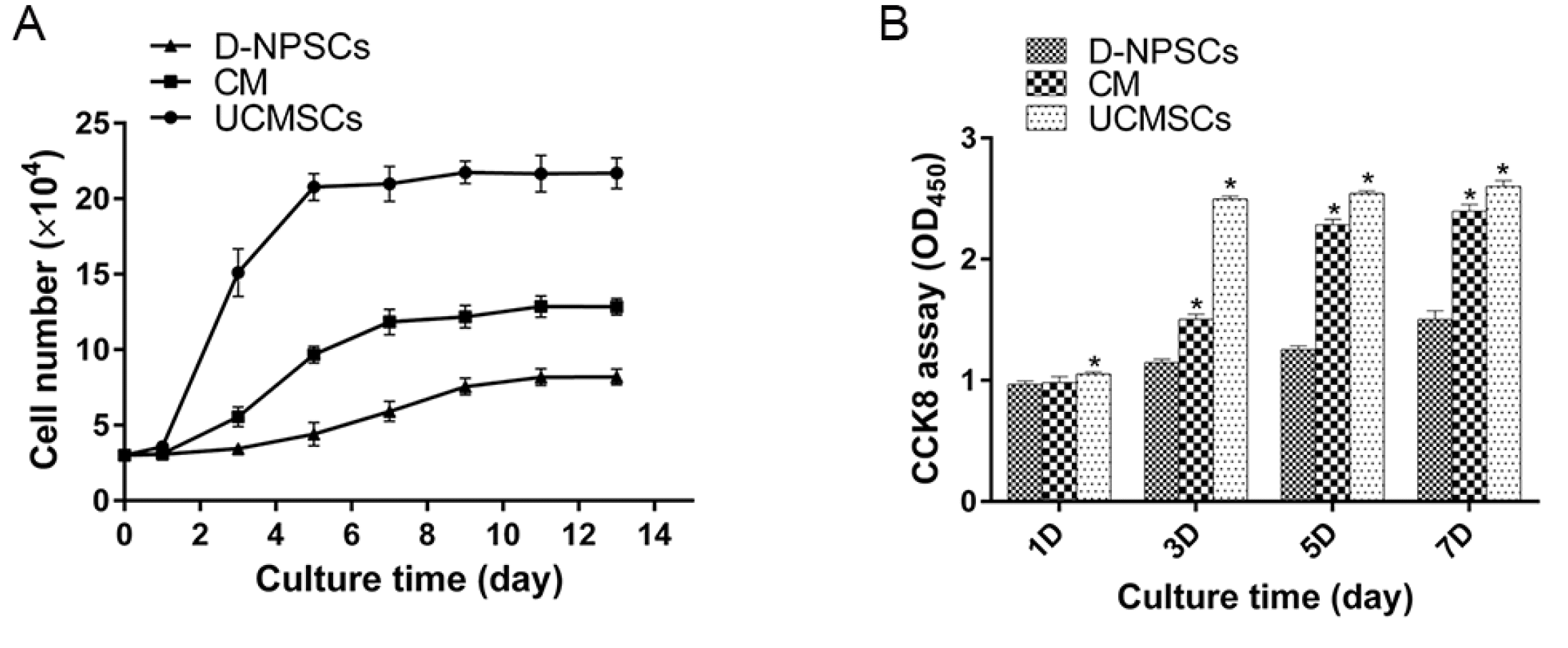
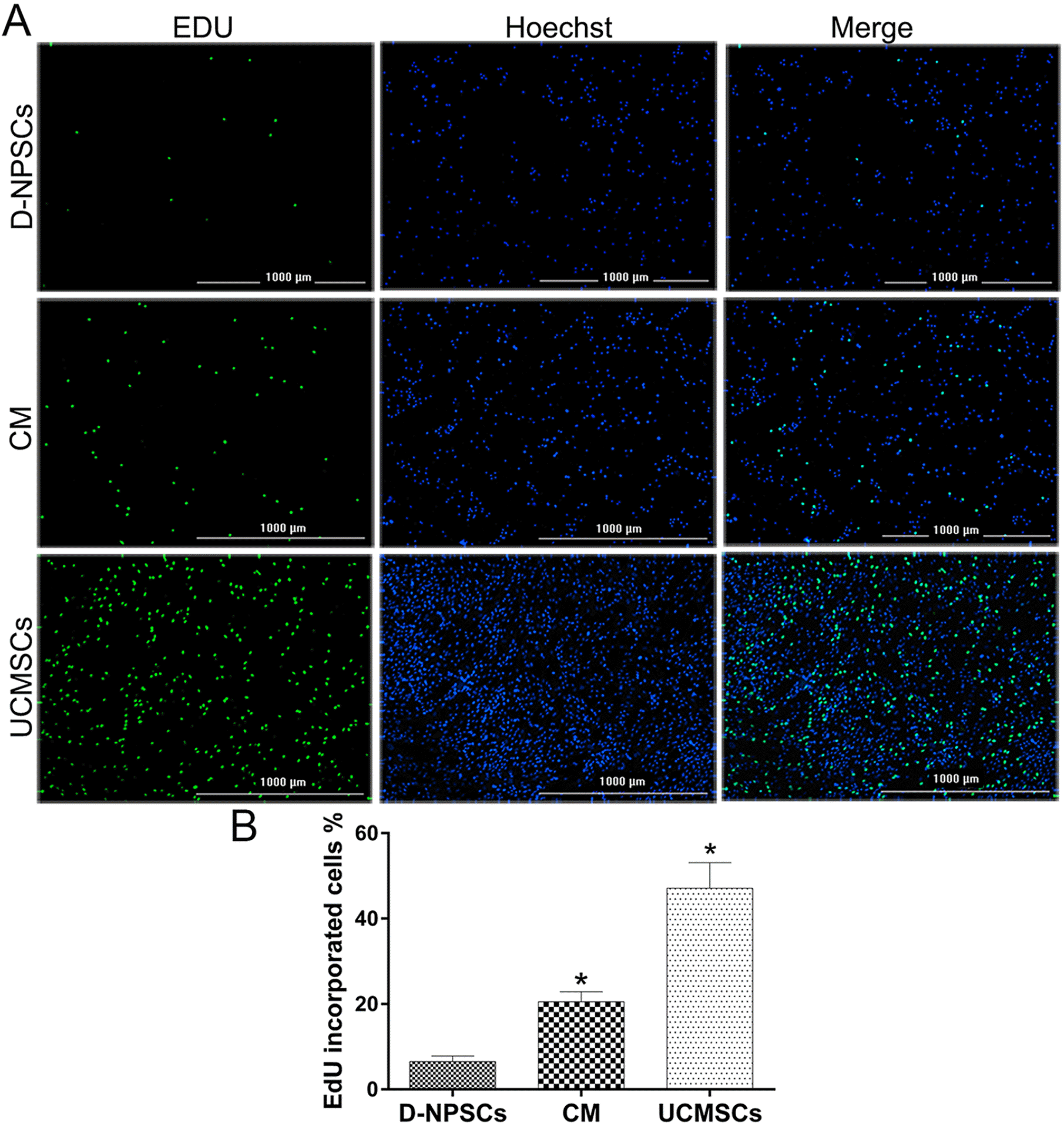
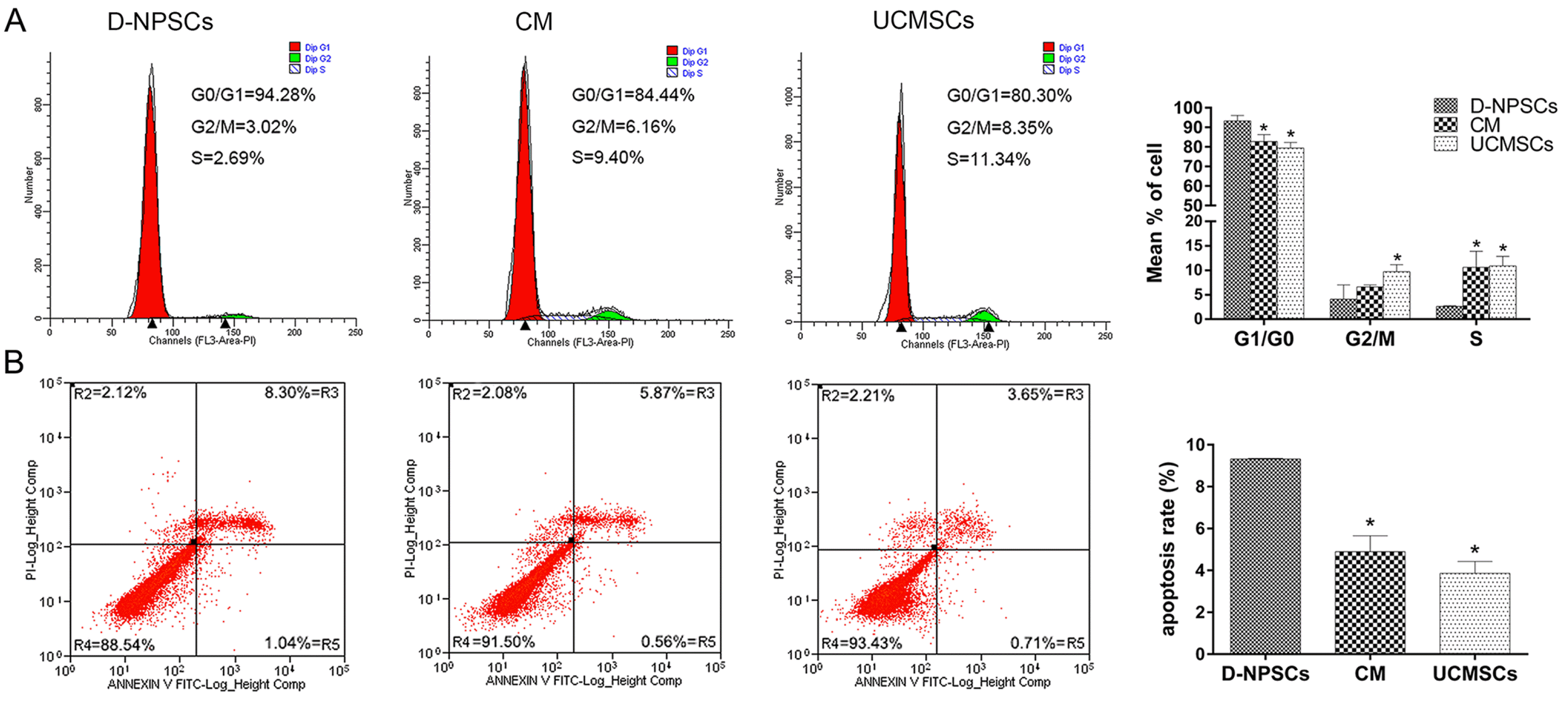




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download