1. Reubinoff BE, Pera MF, Fong CY, Trounson A, Bongso A. 2000; Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat Biotechnol. 18:399–404. DOI:
10.1038/74447. PMID:
10748519.

2. Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. 1998; Embryonic stem cell lines derived from human blastocysts. Science. 282:1145–1147. DOI:
10.1126/science.282.5391.1145. PMID:
9804556.

4. Dahéron L, Opitz SL, Zaehres H, Lensch MW, Andrews PW, Itskovitz-Eldor J, Daley GQ. 2004; LIF/STAT3 signaling fails to maintain self-renewal of human embryonic stem cells. Stem Cells. 22:770–778. DOI:
10.1634/stemcells.22-5-770. PMID:
15342941.

5. Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Schöler H, Smith A. 1998; Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 95:379–391. DOI:
10.1016/S0092-8674(00)81769-9.

6. Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. 2003; Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 17:126–140. DOI:
10.1101/gad.224503. PMID:
12514105. PMCID:
PMC195970.

7. Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. 2003; Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 113:643–655. DOI:
10.1016/S0092-8674(03)00392-1. PMID:
12787505.

8. Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. 2003; The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 113:631–642. DOI:
10.1016/S0092-8674(03)00393-3. PMID:
12787504.

9. Niwa H, Miyazaki J, Smith AG. 2000; Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 24:372–376. DOI:
10.1038/74199. PMID:
10742100.

12. Aubert J, Dunstan H, Chambers I, Smith A. 2002; Functional gene screening in embryonic stem cells implicates Wnt antagonism in neural differentiation. Nat Biotechnol. 20:1240–1245. DOI:
10.1038/nbt763. PMID:
12447396.

13. Haegele L, Ingold B, Naumann H, Tabatabai G, Ledermann B, Brandner S. 2003; Wnt signalling inhibits neural differentiation of embryonic stem cells by controlling bone morphogenetic protein expression. Mol Cell Neurosci. 24:696–708. DOI:
10.1016/S1044-7431(03)00232-X. PMID:
14664819.

14. Kielman MF, Rindapää M, Gaspar C, van Poppel N, Breukel C, van Leeuwen S, Taketo MM, Roberts S, Smits R, Fodde R. 2002; Apc modulates embryonic stem-cell differentiation by controlling the dosage of beta-catenin signaling. Nat Genet. 32:594–605. DOI:
10.1038/ng1045. PMID:
12426568.

15. Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. 2004; Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 10:55–63. DOI:
10.1038/nm979. PMID:
14702635.

16. Kirby LA, Schott JT, Noble BL, Mendez DC, Caseley PS, Peterson SC, Routledge TJ, Patel NV. 2012; Glycogen synthase kinase 3 (GSK3) inhibitor, SB-216763, promotes pluripotency in mouse embryonic stem cells. PLoS One. 7:e39329. DOI:
10.1371/journal.pone.0039329. PMID:
22745733. PMCID:
PMC3383737.

17. Dravid G, Ye Z, Hammond H, Chen G, Pyle A, Donovan P, Yu X, Cheng L. 2005; Defining the role of Wnt/beta-catenin signaling in the survival, proliferation, and self-renewal of human embryonic stem cells. Stem Cells. 23:1489–1501. DOI:
10.1634/stemcells.2005-0034. PMID:
16002782.

18. Kwon C, Arnold J, Hsiao EC, Taketo MM, Conklin BR, Srivastava D. 2007; Canonical Wnt signaling is a positive regulator of mammalian cardiac progenitors. Proc Natl Acad Sci USA. 104:10894–10899. DOI:
10.1073/pnas.0704044104. PMID:
17576928. PMCID:
PMC1904134.

19. Lindsley RC, Gill JG, Kyba M, Murphy TL, Murphy KM. 2006; Canonical Wnt signaling is required for development of embryonic stem cell-derived mesoderm. Development. 133:3787–3796. DOI:
10.1242/dev.02551. PMID:
16943279.

20. Naito AT, Shiojima I, Akazawa H, Hidaka K, Morisaki T, Kikuchi A, Komuro I. 2006; Developmental stage-specific biphasic roles of Wnt/beta-catenin signaling in cardiomyogenesis and hematopoiesis. Proc Natl Acad Sci USA. 103:19812–19817. DOI:
10.1073/pnas.0605768103. PMID:
17170140. PMCID:
PMC1750922.

21. Otero JJ, Fu W, Kan L, Cuadra AE, Kessler JA. 2004; Beta-catenin signaling is required for neural differentiation of embryonic stem cells. Development. 131:3545–3557. DOI:
10.1242/dev.01218. PMID:
15262888.

22. Kim H, Kim S, Song Y, Kim W, Ying QL, Jho EH. 2015; Dual function of Wnt signaling during neuronal differentiation of mouse embryonic stem cells. Stem Cells Int. 2015:459301. DOI:
10.1155/2015/459301. PMID:
25945099. PMCID:
PMC4402205.

24. Arce L, Yokoyama NN, Waterman ML. 2006; Diversity of LEF/TCF action in development and disease. Oncogene. 25:7492–7504. DOI:
10.1038/sj.onc.1210056. PMID:
17143293.

25. Hoppler S, Kavanagh CL. 2007; Wnt signalling: variety at the core. J Cell Sci. 120(Pt 3):385–393. DOI:
10.1242/jcs.03363. PMID:
17251379.

27. Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. 2008; Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 22:746–755. DOI:
10.1101/gad.1642408. PMID:
18347094. PMCID:
PMC2275428.

28. Pereira L, Yi F, Merrill BJ. 2006; Repression of Nanog gene transcription by Tcf3 limits embryonic stem cell self-renewal. Mol Cell Biol. 26:7479–7491. DOI:
10.1128/MCB.00368-06. PMID:
16894029. PMCID:
PMC1636872.

29. Tam WL, Lim CY, Han J, Zhang J, Ang YS, Ng HH, Yang H, Lim B. 2008; T-cell factor 3 regulates embryonic stem cell pluripotency and self-renewal by the transcriptional control of multiple lineage pathways. Stem Cells. 26:2019–2031. DOI:
10.1634/stemcells.2007-1115. PMID:
18467660. PMCID:
PMC2692055.

30. Yi F, Pereira L, Merrill BJ. 2008; Tcf3 functions as a steady-state limiter of transcriptional programs of mouse embryonic stem cell self-renewal. Stem Cells. 26:1951–1960. DOI:
10.1634/stemcells.2008-0229. PMID:
18483421. PMCID:
PMC2743928.

31. Zhang Y, Zhu Z, Ding H, Wan S, Zhang X, Li Y, Ji J, Wang X, Zhang M, Ye SD. 2020; β-catenin stimulates Tcf7l1 degradation through recruitment of casein kinase 2 in mouse embryonic stem cells. Biochem Biophys Res Commun. 524:280–287. DOI:
10.1016/j.bbrc.2020.01.074. PMID:
31987502.

32. Ye S, Zhang T, Tong C, Zhou X, He K, Ban Q, Liu D, Ying QL. 2017; Depletion of Tcf3 and Lef1 maintains mouse embryonic stem cell self-renewal. Biol Open. 6:511–517. DOI:
10.1242/bio.022426. PMID:
28288968. PMCID:
PMC5399551.

33. Ying QL, Stavridis M, Griffiths D, Li M, Smith A. 2003; Convertsion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol. 21:183–186. DOI:
10.1038/nbt780. PMID:
12524553.

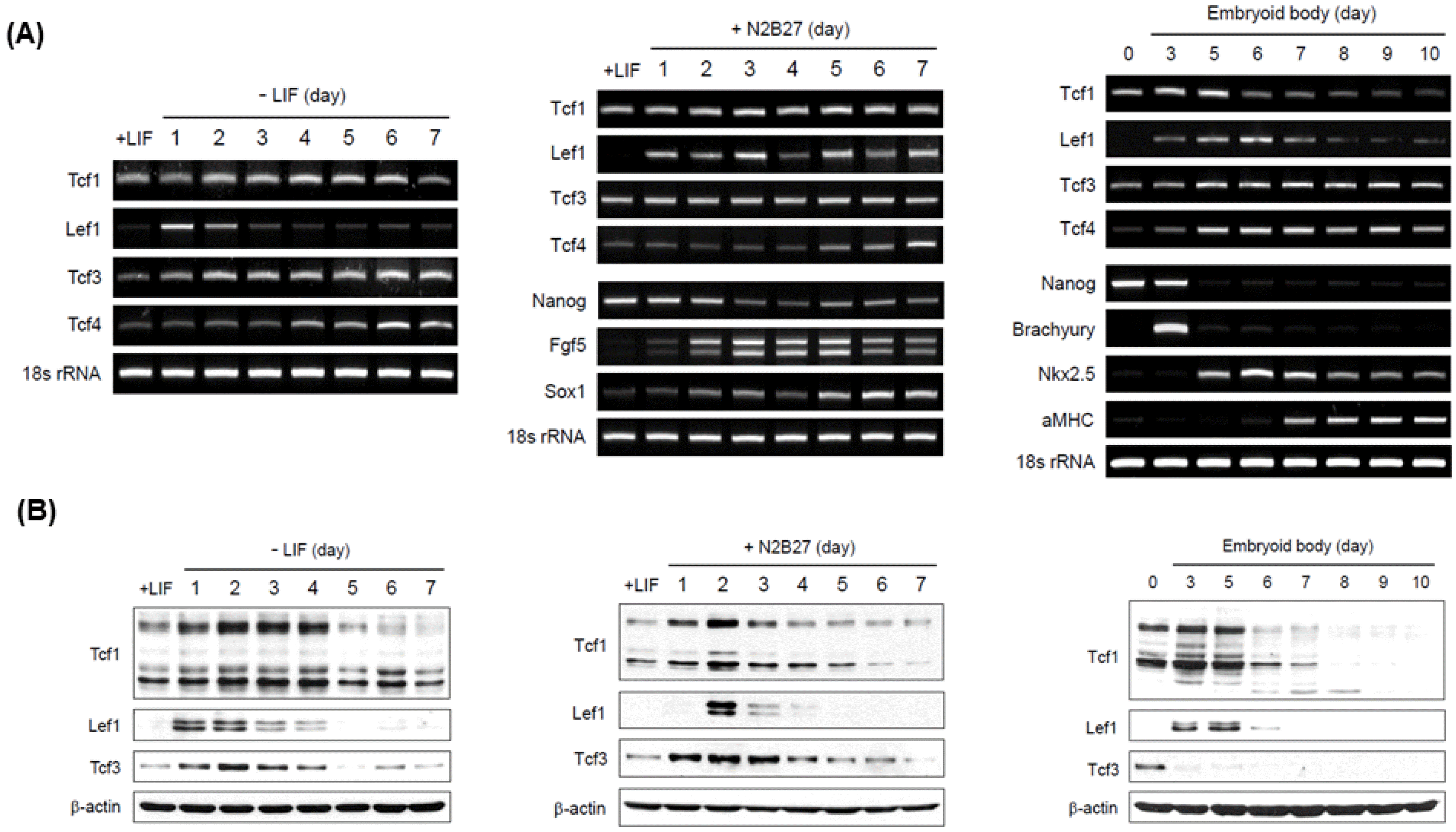
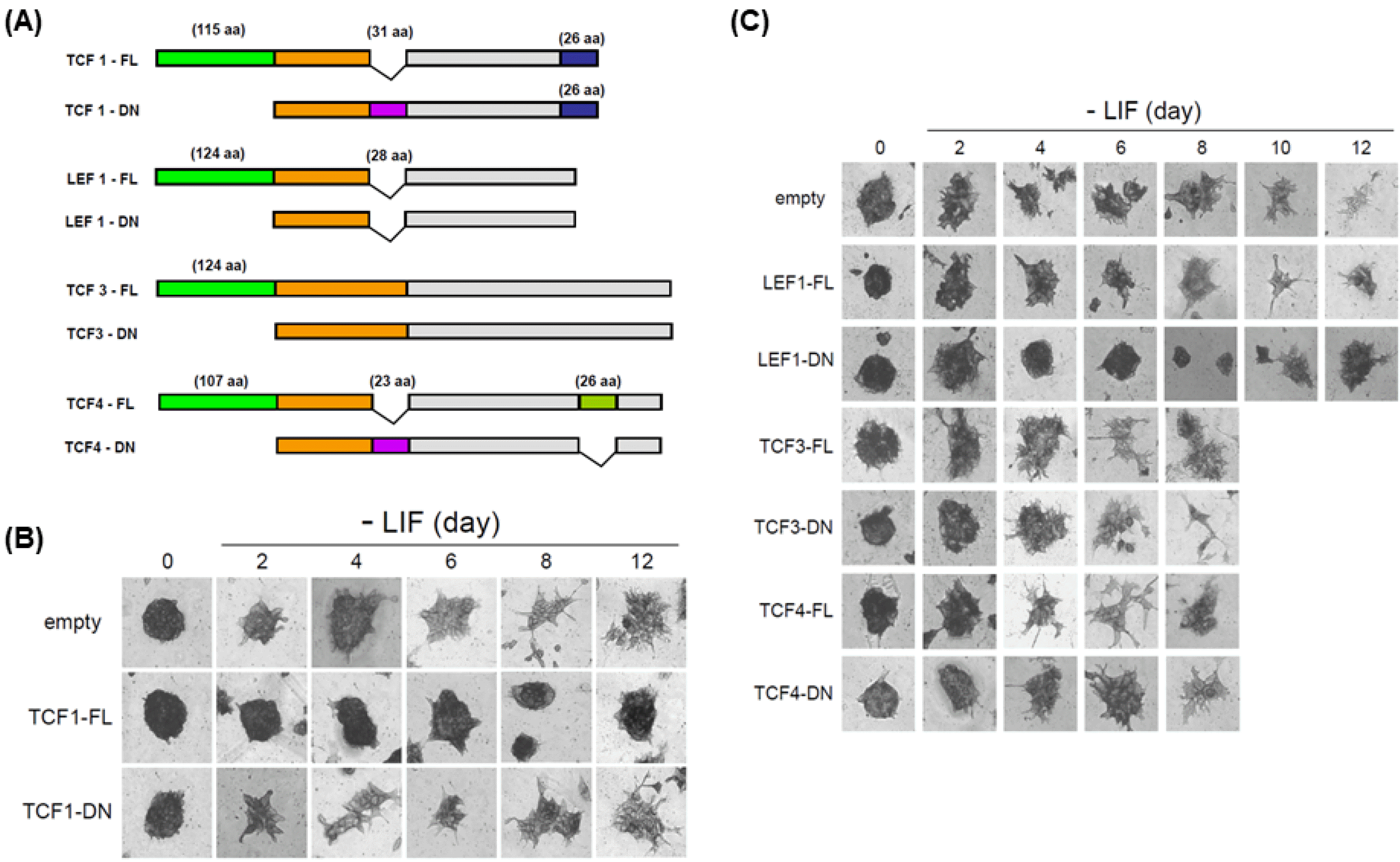
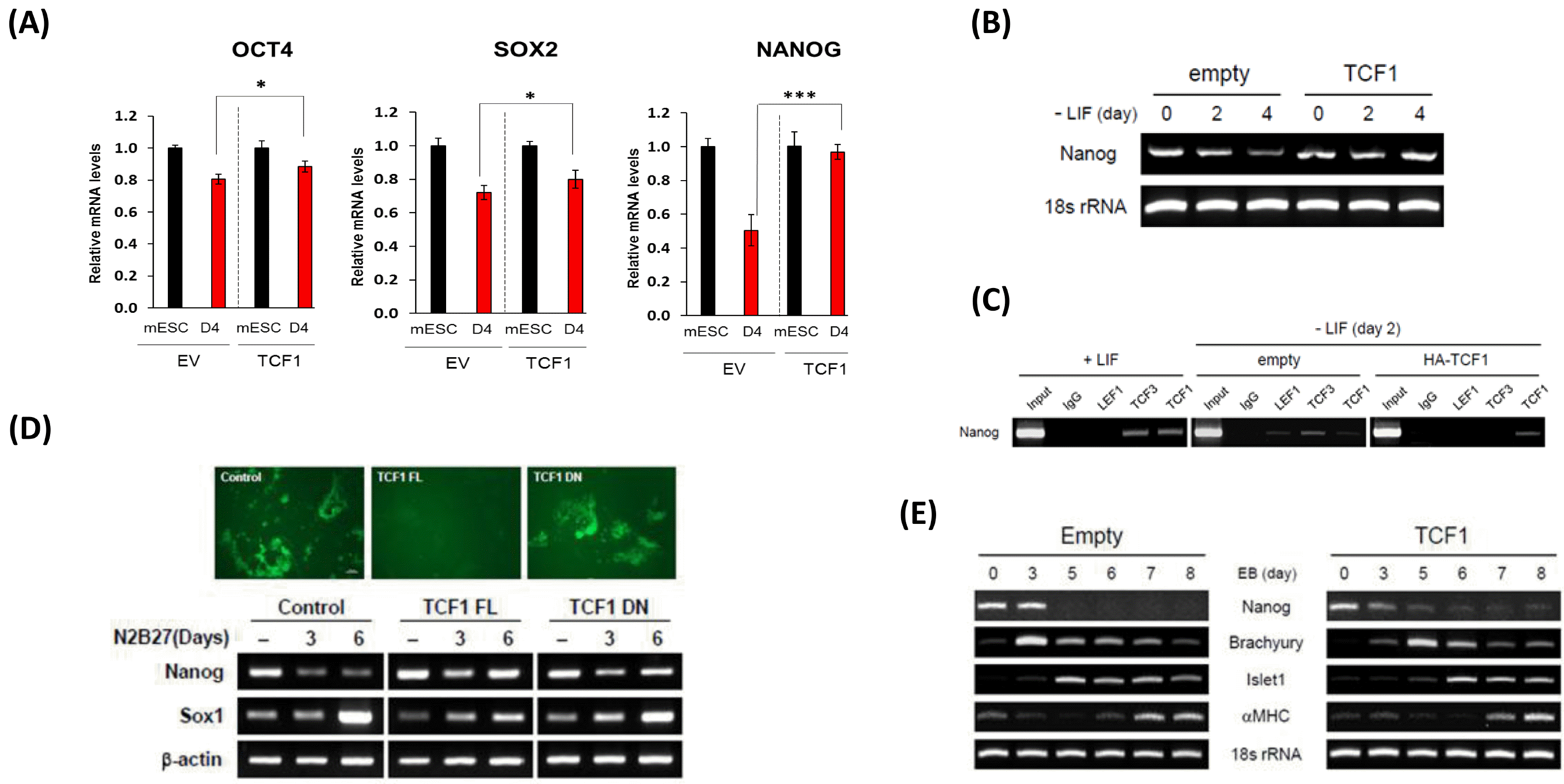
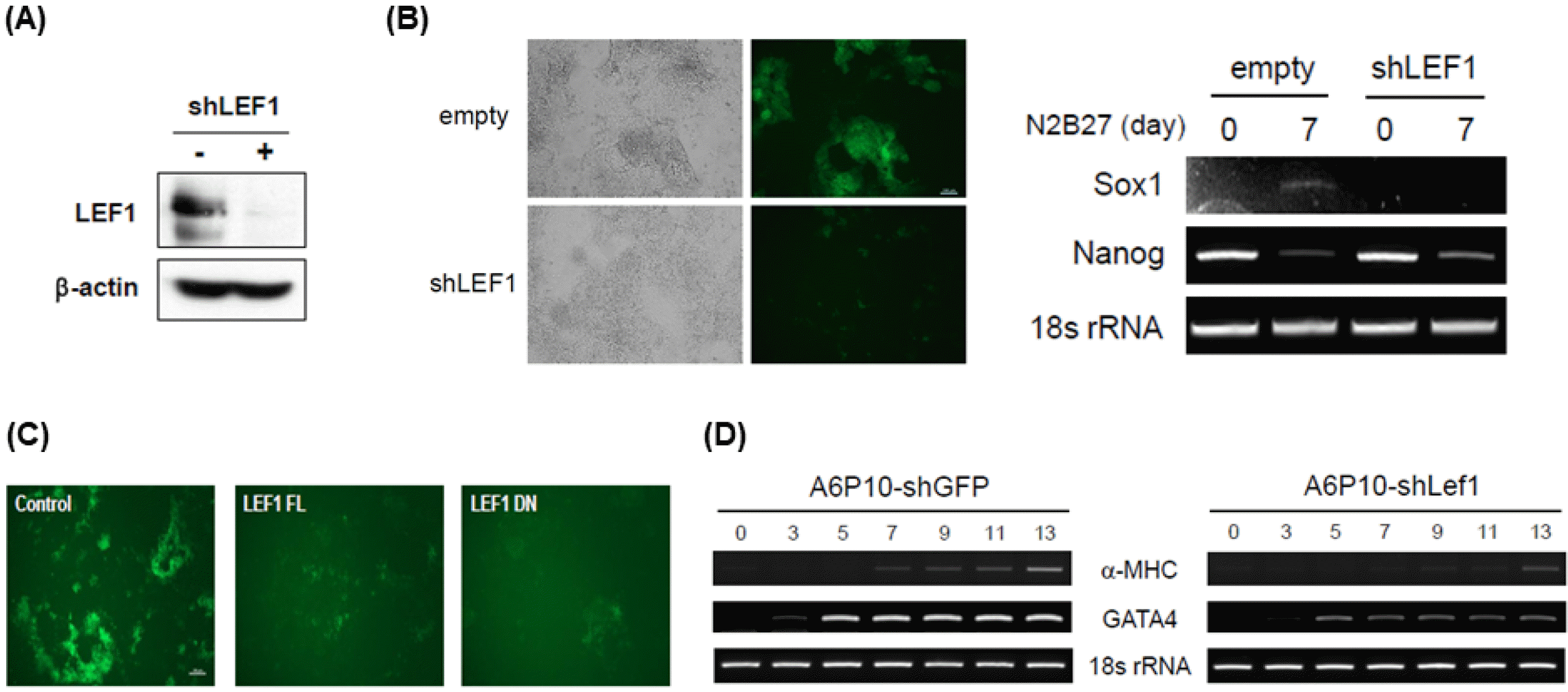




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download