Abstract
Objectives
This study was conducted to develop a generalizable annotation tool for bilingual complex clinical text annotation, which led to the design and development of a clinical text annotation tool, ANNO.
Methods
We designed ANNO to enable human annotators to support the annotation of information in clinical documents efficiently and accurately. First, annotations for different classes (word or phrase types) can be tagged according to the type of word using the dictionary function. In addition, it is possible to evaluate and reconcile differences by comparing annotation results between human annotators. Moreover, if the regular expression set for each class is updated during annotation, it is automatically reflected in the new document. The regular expression set created by human annotators is designed such that a word tagged once is automatically labeled in new documents.
Results
Because ANNO is a Docker-based web application, users can use it freely without being subjected to dependency issues. Human annotators can share their annotation markups as regular expression sets with a dictionary structure, and they can cross-check their annotated corpora with each other. The dictionary-based regular expression sharing function, cross-check function for each annotator, and standardized input (Microsoft Excel) and output (extensible markup language [XML]) formats are the main features of ANNO.
Go to : 
Medical records, such as doctor’s notes and nursing records, must be checked to ensure the quality of clinical research. It is difficult to fully understand patients’ various clinical situations and decision-making using only the available coded information. Clinical documents have a variety of purposes, such as recording significant observations or impressions, treatment plans or activities related to patient treatment, supporting communication and collaboration among medical staff, justification of payment claims, and use as legal records [1]. The unstructured and narrative characteristics of clinical documents are an efficient way for medical staff to make decisions.
However, these characteristics present obstacles for data recycling, systems integration, model development using artificial intelligence (AI), and designing clinical decision support systems. Therefore, efforts are being made to extract information from clinical documents in a structured form for multiple purposes. In particular, as clinical research using machine learning (ML) and AI is rapidly expanding, there is a demand for large-scale clinical data to train these models. Because the amount of data required to train an ML model is substantial, the task of annotation is large and time-consuming [2]. Furthermore, due to the sensitivity of clinical documents, personal information must be removed for use in research. Therefore, a system annotating large-scale data is essential to extract information or remove personal information.
Several clinical document annotation tools have already been developed to support the extraction of machine-readable data from clinical documents. The previously developed Healthcare Data Extraction and Analysis (HEDEA) tool is a Python-based tool for extracting structured information from various clinical documents [3]. It supports the processing of multi-center clinical documents based on regular expressions and enables data integration by centrally identifying patients. The annotator designed by Cedeno Moreno and Vargas-Lombardo [4] uses natural language processing to process words and then maps them to an ontology to infer information. As a text span annotation tool, YEDDA [5] emphasizes the manager’s role in improving post-annotation data quality, especially in the annotation process. Anafora [6] is designed based on a general-purpose cross-platform deployment, which overcomes the limitations of existing local applications. Many existing annotation tools [7–9] have been developed, each of which has its advantages and disadvantages.
However, in Korea, there has been no application for annotating clinical documents in which Korean and English could be mixed in different ways. In addition, most existing tools are system-dependent, biased toward a specific purpose, or resource-intensive, making it difficult to use them for various types of tasks. Therefore, to solve this problem, we developed ANNO, a general annotation tool for information extraction from bilingual clinical notes.
Go to : 
The main purpose of ANNO is to support the tagging of words needed by researchers in clinical documents containing various languages and groups of expressions to create a gold standard set of AI-based predictive models using methods such as ML and deep learning. In fact, performing manual annotation is highly time-consuming and can result in many errors. ANNO has been designed with several major features to help perform these tasks. First, it was designed to have a simple and user-friendly web interface so that human curators, who are the main users of annotation tools, can access it easily. In addition, the type of word or phrase to be annotated (“class”) can be freely configured as required by the user. A user-created class (attribute) and the corresponding regular expression set (value) can be reused by other users, thereby reducing the amount of work needed.
To support stable and ongoing annotation work, we designed a function to store word combinations for each class and manage them by user accounts. Considering that two or more workers cross-validate annotations to correct errors, usually through account management, the color of each worker’s tag is different, which enables the annotation results of one worker to be easily compared with those of another. When a discrepancy occurs, it is highlighted.
In addition, the input file format was designed to use Excel-formatted files by default, so that the corpus can be set to receive various text document structures from each institution. If the row is a clinical document for each patient and an Excel file containing the text to be processed is uploaded into a column, the operator can set the corresponding corpus as the target by entering the column number in which the text to be processed exists. For clinical documents with different structures at different institutions, ANNO can be used freely when manual decisions are made regarding column(s) of the Excel file where the text should be parsed.
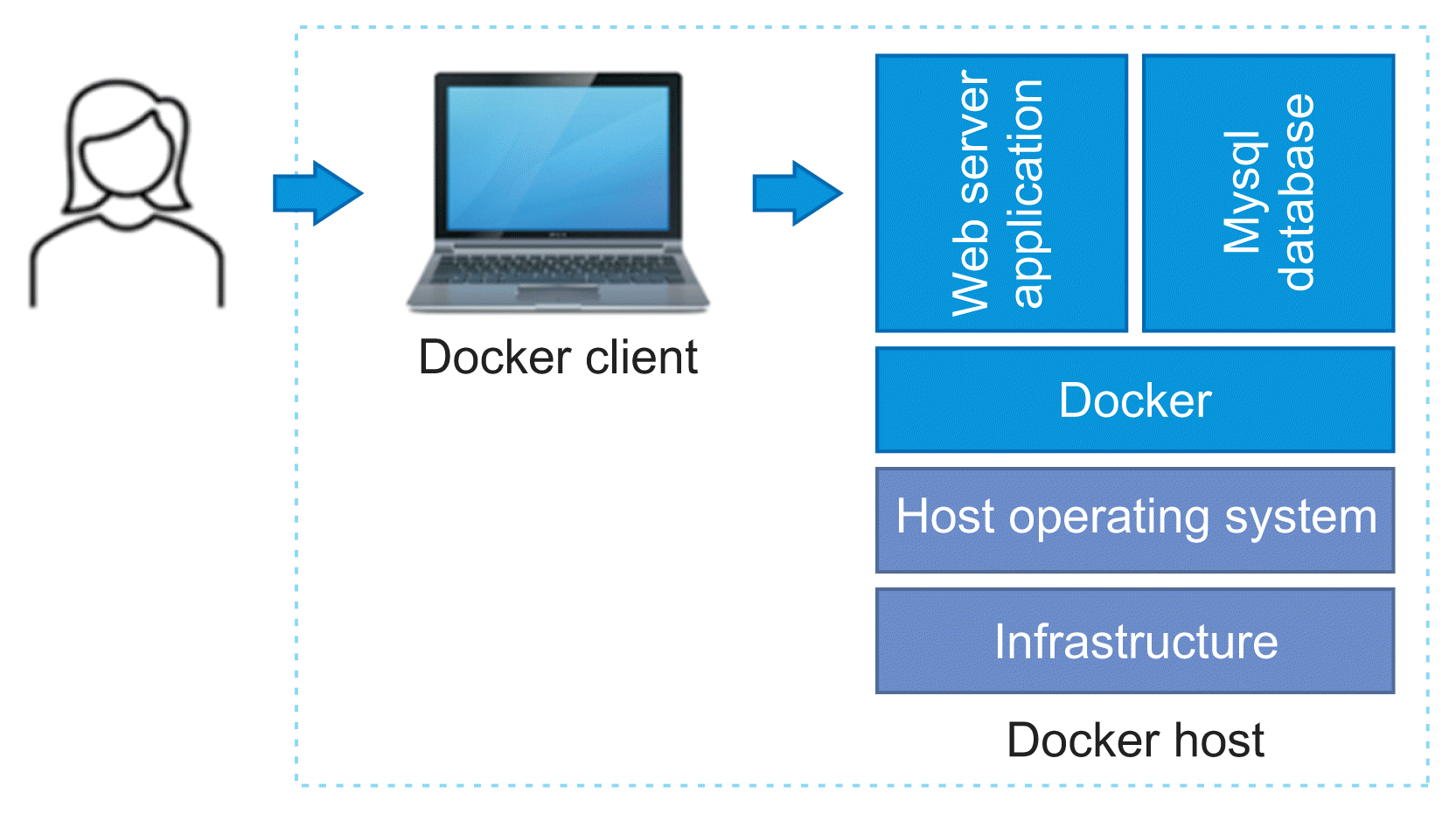
Because ANNO was developed based on a Docker container [10], it can be installed and used stably regardless of the operating system. Docker is a set of platforms as a service that uses operation system-level virtualization to deliver software in packages called containers. By using Docker, the deployment and management of programs can be simplified by abstracting various programs and execution environments into containers and providing the same interface.
Finally, ANNO supports UTF-8 (Unicode Transformation Format-8 bit) encoding and is designed to enable free annotation of clinical documents in which Korean, English, numbers, abbreviations, and special characters are mixed. The output of ANNO is an XML (extensible markup language)-based format, which contains information on the result class annotated with the main corpus, the corresponding word, and the start and end positions of the word. This standardized format supports data exchange and broad compatibility. For a test run, we used several clinical documents from the Seoul National University Hospital (IRB No. H-2001-053-1093).
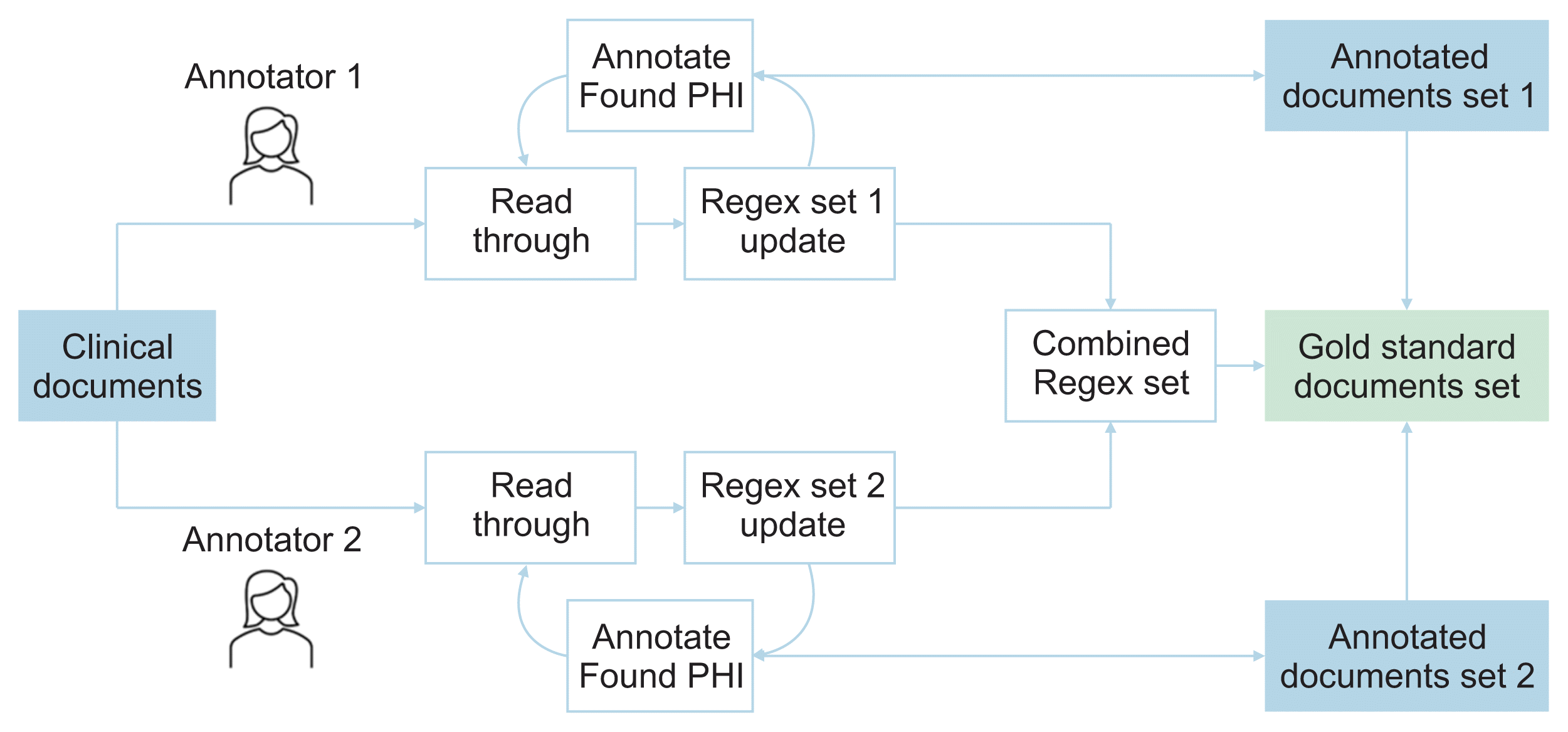
As described above, ANNO is designed such that two or more annotators each annotate and compare the results for each class. If the results annotated by one annotator overlap with another, the color coding of the corresponding word or phrase appears differently. When the annotator classifies a specific word found while reading the corpus into the corresponding class and updates the regular expression set, the corpus is searched using this updated markup set. The results are highlighted and displayed in output form. Finally, each annotator creates a list of files or documents that he or she has reviewed, the regular expression dictionary created while reviewing, and the annotated result file. To enable cross-checking, we created a third-party annotator review scenario. At this time, the third-party reviewer retrieves the input file from the regular expression dictionary created by two different annotators, checks the intersection, difference, and union results of the sets made by two people for each corpus, and corrects or accepts them to create the final gold-standard set (Figure 1).
Software development can be challenging. There is a dependency on the operating system environment, several packages and libraries, and other software components that are required for software functionality. Installation, execution, and maintenance while keeping the software stack up-to-date are complex issues. Previously, virtual machines were used to solve this problem. However, a problem was that virtual machines used excessive resources of the main machine. Although cloud computing can be chosen instead, it can be inefficient for very light applications. In addition, within the Korean medical community, there are concerns about the danger of uploading patients’ personal information to the cloud. We built ANNO using Docker to solve this problem. With Docker containers, the operating system and applications are separated and dependencies on other applications are removed; therefore, these variables need not be considered in software development and maintenance. Applications can be run stably in different computing environments, and rapid development and deployment are possible regardless of the development environment. Figure 2 shows the system architecture of ANNO developed using Docker.
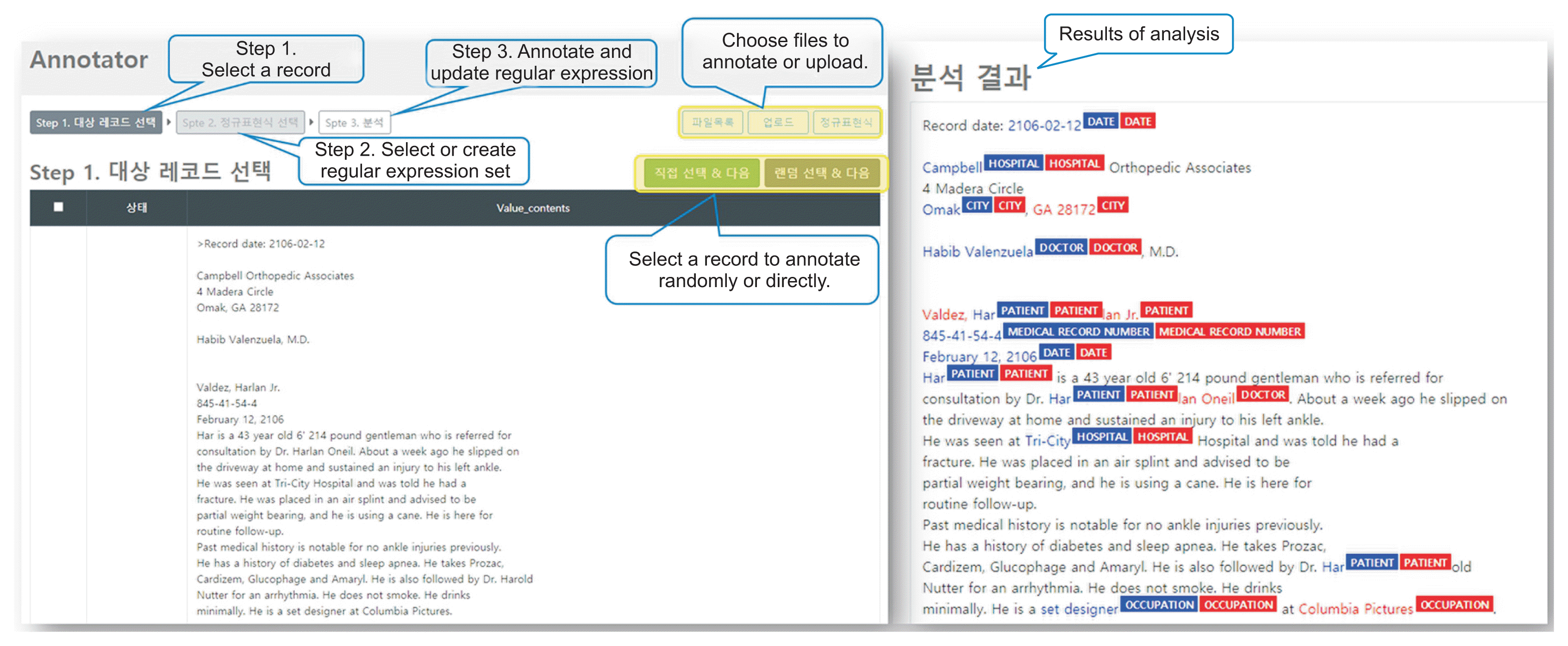
ANNO was designed with a thorough consideration of the convenience of the annotators. ANNO is accessed via the annotator’s account, and functions such as class definitions of what information should be extracted, stopping while annotating, and modifying the regular expression set can all be performed by the annotator through the web. Individually created regular expression sets are private; however, one can choose to keep them private or provide access to other users. When inputting a file in Excel format, one can select whether to perform operations sequentially from step 1 or annotate through random sampling, check through the preview, and manually select data to annotate one by one. The user interface and sample data for the first selection step are shown in the upper portion of Figure 3. To develop a function that can freely annotate words or phrases in various clinical documents and a function that supports cross-validation between human annotators, we extracted data through SUPPREME, a clinical data warehouse at Seoul National University Hospital. Five types of clinical documents were used: first visits for hospitalization, discharge records, surgery records, outpatient first visits, and emergency records. After selecting a record, if there is an existing regular expression set, the operator can either import it and continue the operation or create a new set and use it. Annotation results help to assist the annotator by color-coding the words found in the original document, as shown in the lower panel of Figure 3. The found word is highlighted in blue, and the corresponding class type is displayed as a colored box. Matching operations performed by different annotators are displayed in red. In Figure 3, we used sample clinical text.
Figure 4 shows the resultant file created after the annotation. This file conforms to the XML standard encoded in UTF-8. By arranging the original text and the annotated result text together, the predictive model can be verified immediately with one file while also performing additional tasks, such as employing the use of an AI model. It is also possible to check the class of the annotated result using tags, the first and last words in a phrase, and the meaning of the actual words.
To test the performance of ANNO, we annotated some clinical documents to detect personal identifiers. Five types of personal identifiers (patient name, doctor name, hospital name, department name, and location) were annotated using ANNO for 1,211 Seoul National University Hospital admission notes. The input was an Excel file, and the annotation result was saved in the XML file format. Fifty-eight of the 1,211 items were excluded because an error occurred when annotating them using ANNO, and the remaining 1,135 items were annotated successfully. An analysis of the 58 annotation errors showed that 29 cases (50.0%) were hospital names which were a combination of a local name and a hospital name, 16 cases (27.5%) were department names given as an abbreviation in English, nine cases (15.5%) occurred when hospital names contained person names, and four cases (6.8%) were other cases. Among the 1,135 hospitalization records annotated without error, we identified a total of 145 hospital names, 136 department names, 173 doctor names, 31 regional names, and 15 patients’ names, which were successfully annotated.
Go to : 
This paper introduced an annotation system to support the development of AI models that can perform annotation for each class and operator in free-text clinical documents and continuously compare and analyze them. We propose a rule-based approach to dealing with complex document structures and the diversity that exists between different organizations, which can be done by inserting a set of keywords or making updates as the curators conduct reviews. This system allows freely setting classes, as well as sharing of classes and regular expression templates belonging to the class, thereby efficiently extracting data for each purpose. The input file format is an Excel file, which is widely used in hospital research data warehouses. The output file format is a standardized and structured XML file format, which improves ease of use.
In addition, the accuracy of the gold standard set construction could be improved through the color-coding function to check mismatches of the annotation results of various curators to build the gold standard set. Finally, it was possible to install all host and client packages in a Docker container without any operating system dependencies. In conclusion, this system can be used as a useful program for various institutions and individual researchers to establish a gold standard set for de-identification and information extraction of clinical documents, which are increasingly demanded for ML and AI model development.
Go to : 
Acknowledgments
This research was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (No. HR21C0198).
Go to : 
References
1. Spasic I, Nenadic G. Clinical text data in machine learning: systematic review. JMIR Med Inform. 2020; 8(3):e17984.

2. Halevy A, Norvig P, Pereira F. The unreasonable effectiveness of data. IEEE Intell Syst. 2009; 24(2):8–12.

3. Aggarwal A, Garhwal S, Kumar A. HEDEA: a Python tool for extracting and analysing semi-structured information from medical records. Healthc Inform Res. 2018; 24(2):148–53.

4. Cedeno Moreno D, Vargas-Lombardo M. Design and construction of a NLP based knowledge extraction methodology in the medical domain applied to clinical information. Healthc Inform Res. 2018; 24(4):376–80.

5. Yang J, Zhang Y, Li L, Li X. YEDDA: a lightweight collaborative text span annotation tool [Internet]. Ithaca (NY): arXiv.org;2017. [cited at 2022 Jan 19]. Available from: https://arxiv.org/abs/1711.03759
.
6. Chen WT, Styler W. Anafora: a web-based general purpose annotation tool. Proc Conf. 2013; 2013:14–9.
7. Bontcheva K, Cunningham H, Roberts I, Roberts A, Tablan V, Aswani N, et al. GATE Teamware: a web-based, collaborative text annotation framework. Lang Resour Eval. 2013; 47(4):1007–29.

8. Lenzi VB, Moretti G, Sprugnoli R. CAT: the CELCT annotation tool. In : Proceedings of the 8th International Conference on Language Resources and Evaluation (LREC); 2012 May 23–25; Istanbul, Turkey. p. 333–8.
9. Ogren P. Knowtator: a protégé plug-in for annotated corpus construction. In : Proceedings of the Human Language Technology Conference of the NAACL; 2006 Jun 4–9; New York, NY. p. 273–5.
10. Merkel D. Docker: lightweight Linux containers for consistent development and deployment. Linux J. 2014; 2014(239):2.
Go to : 




 PDF
PDF Citation
Citation Print
Print




 XML Download
XML Download