Abstract
Combined hepatocellular-cholangiocarcinoma (cHCC-CCA) is a malignant primary liver carcinoma characterized by the unequivocal presence of both hepatocytic and cholangiocytic differentiation within the same tumor. Recent research has highlighted that cHCC-CCAs are more heterogeneous than previously expected. In the updated consensus terminology and WHO 2019 classification, “classical type” and “subtypes with stem-cell features” of the WHO 2010 classification are no longer recommended. Instead, it is recommended that the presence and percentages of various histopathologic components and stem-cell features be mentioned in the pathologic report. The new terminology and classification enable the exchange of clearer and more objective information about cHCC-CCAs, facilitating multi-center and multi-national research. However, there are limitations to the diagnosis of cHCC-CCA by imaging and biopsy. cHCC-CCAs showing typical imaging findings of HCC could be misdiagnosed as HCC and subjected to inappropriate treatment, if other clinical findings are not sufficiently considered. cHCC-CCAs showing at least one of the CCA-like imaging features or unusual clinical features should be subjected to biopsy. There may be a sampling error for the biopsy diagnosis of cHCC-CCA. An optimized diagnostic algorithm integrating clinical, radiological, and histopathologic information of biopsy is required to resolve these diagnostic pitfalls.
Combined hepatocellular-cholangiocarcinoma (cHCC-CCA) is a primary liver cancer (PLC) showing both hepatocytic and cholangiocytic differentiation, it reportedly accounts for about 1–5% of all PLCs.1–3 Given the heterogeneous histopathologic nature of cHCC-CCAs, imaging or biopsy diagnosis is challenging, and the ideal diagnostic strategy for cHCC-CCAs in non-surgical candidates is not sufficiently well defined. Despite several recent investigations, the pathogenesis, histopathology, and genetics of this enigmatic tumor have not been fully understood. Reflecting on the current understanding, the consensus terminology of cHCC-CCA was published in 2018, and the WHO classification of cHCC-CCA was revised in 2019.1,4 Herein, we aimed to comprehensively review the updated information on the pathologic and radiologic diagnosis of cHCC-CCAs.
In the updated 2018 consensus terminology and WHO 2019 classification,1,4 cHCC-CCA is defined as a PLC with the unequivocal presence of both hepatocytic and cholangiocytic differentiation within the same tumor, similar to WHO 2010 classification. cHCC-CCA is characterized by complex morphological and immunophenotypical features and can be diagnosed regardless of the percentage of each component if the components are present unequivocally. However, multi-focal HCC or CCA, collision of HCC and CCA arising separately, any form of hepatoblastoma or variants, pediatric transitional liver cell tumor or variants, and HCC or CCA with neuroendocrine components should not be classified as cHCC-CCAs.
In the WHO 2010 classification, cHCC-CCAs were categorized as “classical type” and three subtypes with stem-cell features, including “typical subtype,” “intermediate-cell subtype,” and “cholangiolocellular subtype”.5 However, criticisms have been raised about the stem cell subtypes. Tumor cells with stem/progenitor cell features can be observed in most cHCC-CCAs, including the classical type. Besides, some PLCs can display two or more histologic components and often do not match the specific subtype of the WHO 2010 classification.6,7 Therefore, in the updated WHO 2019 classification, the use of “cHCC-CCA with stem-cell feature” is no longer recommended.8 It is recommended that the presence and percentages of various histopathologic components and stem-cell features be mentioned in the pathologic report.
Cholangiolocarcinoma (CLC) comprises tumor cells resembling cholangioles (canals of Hering), where the hepatic stem/progenitor cells are located.9,10 CLC also shows enriched CCA-like features and the absence of HCC-like features in the molecular profile, which is distinct from the phenotypes of other stem-cell subtypes of cHCC-CCA.11 However, recent morphometric and immunohistochemical (IHC) analyses revealed that the CLC originated not from the cholangiole but the interlobular bile duct.12 In the updated WHO 2019 classification, the CLC is classified as cHCC-CCA only when it is mixed with HCC or intermediate carcinoma. However, if the CLC component exists alone or is mixed with CCA only, it is classified as CCA, not cHCC-CCA.
Intermediate cell carcinoma is composed of tumor cells showing intermediate features between hepatocytes and cholangiocytes at the cellular level, and it displays both hepatocytic and cholangiocytic IHC markers. PLCs comprising only intermediate cells are diagnosed as intermediate cell carcinoma, and those showing mixed intermediate cell carcinoma and other types of tumors (HCC, CCA, or CLC) are classified as cHCC-CCAs. Further studies are needed to clarify whether intermediate cell carcinoma is a distinct clinicopathological entity rather than a histopathological spectrum.
The diagnosis of cHCC-CCA should be based on the histomorphology on hematoxylin-eosin stain. There are occasions when it is not easy to assess the HCC or CCA area based on histomorphology, especially when the tumor is poorly differentiated, and in such case, immunohistochemistry can be useful to confirm hepatocytic and/or cholangiocytic differentiation. However, the expression of IHC markers alone, without the corresponding histomorphologic features, is not adequate for diagnosis. IHC markers for hepatocytic differentiation include HepPar-1 (75–85% positivity), Arginase-1 (85–95% positivity), polyclonal carcinoembryonic antigen (CEA) with canalicular expression (50–80% positivity), CD10 with canalicular expression (50–75% positivity), and alpha-fetoprotein (AFP) (30% positivity), among others; IHC markers for cholangiocytic differentiation include K7 (>90% positivity), K19 (>75% positivity) and Ep-CAM (>90% positivity), among others. A wide variety of IHC markers, including K19, EpCAM, CD56, KIT, and CD133, have been used to confirm the stem/progenitor cell phenotype. Some of these markers, including K19, EpCAM, and CD56, can also be expressed in cholangiocytes at various development stages. Therefore, it is important that these antibodies be used and interpreted by an experienced pathologist, taking into account the tumor histomorphology.
The pathogenesis of cHCC-CCA remains elusive. Two possibilities have been postulated regarding the pathogenesis of cHCC-CCA: 1) trans-differentiation of HCC or CCA into the other component, and 2) derivation of cHCC-CCA from the hepatic stem/progenitor cell.13 cHCC-CCAs often show tumor cell components with stem-cell features, and thus, liver stem/progenitor cells have been proposed as a potential origin of cHCC-CCAs.14 Recently, increasing clinical and experimental evidence indicated that cHCC-CCA could result from plasticity or the dedifferentiation of PLC. In a mouse model, PLCs from the same cell of origin were suggested to show various hepatocytic/cholangiocytic differentiation and stem-cell features,15 and advanced HCC was reported to show molecular and pathologic features of CCA.16 Among PLCs treated with trans-arterial chemoembolization, the occurrence of cHCC-CCA was reported, suggesting the potential microenvironmental regulation of tumor trans-differentiation. 17
Despite recent active investigations since early 2000, the genetic and molecular features of cHCC-CCA remain unclear. The recently reported molecular features of cHCC-CCAs are summarized in Table 1; these are significantly heterogenous. It should be noted that these studies were performed on a limited series, reflecting the low frequency of this tumor. A genetic study of each microdissected component of cHCC-CCA revealed that HCC and CCA components shared genetic and chromosomal aberrations in most cases, suggesting a single clonal origin.11,18–20 Several studies have reported that the mutation profile of cHCC-CCA is similar to that of CCA.21 In contrast, more recent studies have reported that the mutation profile is rather similar to that of HCC.20,22,23 Moeini et al.11 studied gene expression via unsupervised clustering and reported the molecular features of classical, stem cell, and CLC types. Concerning the background liver, the genome-wide substitution patterns of cHCC-CCAs with chronic hepatitis overlapped with HCCs, whereas those of hepatitis-negative cHCC-CCAs were diverse. 22 These findings suggest that the genetic characteristics of cHCC-CCAs are heterogeneous, similar to their histopathologic features.
As with HCCs, cHCC-CCAs show male predominance. In cHCC-CCA, the incidence of chronic hepatitis and cirrhosis is 23–38% and 50–62%, respectively; these ratios are similar to those of HCC,24–27 or intermediate between those of HCC and CCA.24–31 Previously, a low incidence of chronic hepatitis/cirrhosis was reported in western series;32 however, in recently reported country-wide data from the United States, the incidence of chronic hepatitis/cirrhosis is similar to that in HCC. The etiologies of chronic hepatitis/cirrhosis include hepatitis B (27–80%), hepatitis C (4–38%), or alcoholic liver disease (6–36%).24–31 The chronic hepatitis and cirrhosis are risk factors not only for HCCs but also for CCAs.33 However, the association of cHCC-CCAs with other risk factors for CCAs, including parasitic fluke, primary sclerosing cholangitis, and choledocholithiasis, remains unknown.
Serologic markers, including AFP, CEA, and carbohydrate antigen 19-9 (CA19-9), are often evaluated when a PLC is suspected. In cHCC-CCA, serum AFP was reported to be similar to that in HCC and higher than that in CCA in most reports;29–32,34,35 however, in some reports, the lower AFP levels were reported to be similar to that in CCAs.36 Varying levels of serum CA19-9 in cHCC-CCAs have been reported; these were not significantly higher than those in HCCs.27,36,37 The serum CEA level in cHCC-CCA was not significantly different from that in HCC.27,29,32 Discordant serologic tumor marker levels and imaging findings may be a clue to the diagnosis of cHCC-CCA.38
The primary goal of imaging diagnosis of PLC is to distinguish HCC from other PLCs, including cHCC-CCA and CCA. In high-risk individuals for HCC, typical imaging findings of HCC (arterial enhancement and washout, or LR-5 of Liver Imaging and Reporting and Data System [LI-RADS]) show very high positive predictive value for HCC; therefore, most HCC management guidelines allow imaging diagnosis of HCC without biopsy in those patients.39,40 The definition of high-risk individuals is similar across guidelines with minor differences. Individuals with chronic hepatitis B or cirrhosis of any cause are included in the high-risk category according to most guidelines.39 Most patients with cHCC-CCA can also be included in the high-risk category, as the incidence of chronic hepatitis and cirrhosis in such patients is similar or slightly lower than that in patients with HCC. When PLC is suspected in patients who are not at high risk for HCC, the lesion should be subjected to biopsy, and not imaging diagnosis alone. LI-RADS is an imaging diagnostic algorithm for HCC, integrated into the American Association for the Study of Liver Diseases (AASLD) guideline.40,41 We can categorize the observations into the following LI-RADS categories: LR-1 (definitely benign), LR-2 (probably benign), LR-3 (intermediate probability of malignancy), LR-4 (probably HCC), LR-5 (definitely HCC), and LR-M (probably or definitely malignant, not necessarily HCC). When a hepatic lesion is categorized as LR-5, it can be diagnosed as HCC without a biopsy. When the lesion is categorized as LR-M, a biopsy is recommended for confirmative diagnosis. The computed tomography (CT) and magnetic resonance imaging (MRI) findings of the recently reported cHCC-CCA are summarized in Table 2. The radiologic appearance of cHCC-CCA can resemble either HCC or CCA; therefore, cHCC-CCA is often misdiagnosed as HCC or CCA.42 The arterial enhancement pattern on CT or MRI is one of the most critical imaging features that distinguishes HCC from other PLCs. HCCs often show non-peripheral enhancement in the arterial phase, whereas CCAs often show peripheral (rim-like) enhancement. For cHCC-CCA, the most common arterial phase enhancement pattern is a peripheral enhancement, seen in approximately 39–100% of cHCC-CCAs (Fig. 1, 2).38,42–48 However, approximately 40–59% of cHCC-CCAs exhibit a non-peripheral enhancement pattern in the arterial phase.24,42,43,45,47 The washout and capsular appearance are major features that, in conjunction with non-peripheral enhancement, are suggestive of HCC; they are reported to be observed in 27–67% and 9–28% of cHCC-CCAs, respectively (Fig. 3).24,38,42,43,45,47,48
Although a considerable number of cHCC-CCAs show major features suggesting HCC, the ancillary features suggesting a non-HCC malignancy should also be taken into account to reduce misdiagnosis. These ancillary features, called LR-M features, include targetoid appearance (peripheral arterial enhancement, peripheral washout, and delayed central enhancement), marked diffusion restriction, and infiltrative appearance. Potretzke et al.45 reported that 54% (33/61) of cHCC-CCAs showed major features typical of HCC, but 88% (29/33) of them demonstrated at least one LR-M feature. Lee et al.49 reported that at least one LR-M feature was observed in 94% (31/33) of cHCC-CCAs and 51% (34/66) of HCCs. It was suggested that cHCC-CCA could be diagnosed with a sensitivity of 55% and specificity of 94% when the lesion exhibits three or more LR-M features. Other recent studies reported that 61% of cHCC-CCAs were classified as LR-M and 23–36% as LR-5.47,48 When cHCC-CCAs are categorized as LR-5, they might lead to inappropriate treatment strategies.
HCC-like or CCA-like (LR-M) imaging features do not always reflect the dominant histologic component of cHCC-CCA. HCC-dominant cHCC-CCA might show peripheral arterial enhancement, and CCA-dominant cHCC-CCA might exhibit global enhancement.24,50 Interestingly, several recent reports have proposed that cHCC-CCAs with HCC-like imaging patterns demonstrate a better prognosis than those with a CCA-like imaging pattern. The imaging pattern is a better surrogate than the histologic pattern for predicting outcome after hepatic resection.24,48,50,51 The clinical, histological, and radiological characteristics of PLCs are summarized in Table 3.
As only part of the tumor tissue can be sampled by percutaneous biopsy, there is an effect of sampling on the diagnosis of cHCC-CCA (Fig. 4). For those cases of cHCC-CCA composed predominantly of HCC or CCA area, the other components might not be included in the biopsied tissue, making a sampling error leading to an incorrect diagnosis. Gigante et al.52 studied 21 cases of biopsy-resection-matched cHCC-CCAs. They found that only 48% (10/21) of cHCC-CCAs were diagnosed correctly by biopsy. The remaining 52% (11/21) cHCC-CCAs were misdiagnosed as CCAs or HCCs. The expression of IHC markers often show similar patterns in biopsied and resected tumor tissues, which may help in the diagnosis of cHCC-CCA in biopsy specimens.52,53 Besides, there is little knowledge on whether radiologic imaging can distinguish various histologic components. Even if possible, small tumor areas with different histologic components might be difficult to obtain by image-guided biopsy.
There are still many unknowns in the pathogenesis, histopathology, and genetics of cHCC-CCAs. Recent research has highlighted that cHCC-CCAs are more heterogeneous than previously expected. The recently updated consensus terminology and the WHO 2019 classification for cHCC-CCAs, enabling clear and informative descriptions, are expected to facilitate multi-center and multi-national research.
Most cHCC-CCAs arise in the background of chronic hepatitis or cirrhosis; therefore, the primary differential imaging diagnosis is HCC. cHCC-CCAs showing typical imaging findings of HCC can be misdiagnosed and subjected to inappropriate treatment if other clinical findings are not adequately considered. Using only imaging modalities may lead to misdiagnosis of cHCC-CCAs with minor histologic components. Nevertheless, several reports have demonstrated the prognostic significance of imaging findings of cHCC-CCA, suggesting that imaging is a useful clinical decision-making tool at the preoperative stage.
As most cHCC-CCAs show at least one LR-M (CCA-like) imaging feature in addition to those of HCC, it is often subjected to biopsy for diagnosis. However, there may be sampling error, as only a part of the tumor tissue is sampled during biopsy. For appropriate pathologic diagnosis of cHCC-CCAs, surgical specimens including all tumor components, even minor ones, are preferred. It is necessary to develop an optimal diagnostic algorithm using clinical, radiologic, and histopathologic biopsy information. Several retrospective single-center studies attempted to optimize radiological criteria utilizing LR-M features, combining radiologic and serologic findings, or combining radiological findings and biopsy results.38,49,52 Because these criteria do not have sufficient diagnostic accuracy for cHCC-CCAs, further optimization and external validation are required. Furthermore, the discovery of non-invasive molecular surrogates, such as novel serum markers, could be a solution for overcoming various obstacles in the diagnosis of cHCC-CCA.
Notes
REFERENCES
1. Brunt E, Aishima S, Clavien PA, Fowler K, Goodman Z, Gores G, et al. cHCC-CCA: Consensus terminology for primary liver carcinomas with both hepatocytic and cholangiocytic differentation. Hepatology. 2018; 68:113–126.
2. Edmondson HA, Steiner PE. Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer. 1954; 7:462–503.
3. Wachtel MS, Zhang Y, Xu T, Chiriva-Internati M, Frezza EE. Combined hepatocellular cholangiocarcinomas; analysis of a large database. Clin Med Pathol. 2008; 1:43–47.
4. Sempoux C, Kakar S, Kondo F, Schirmacher P. Combined hepatocellular-cholangiocarcinoma and undifferentiated primary liver carcinoma. World Health Organization. WHO Classification of Tumours. 1:5th ed. 2019. p. 260–262.
5. Gera S, Ettel M, Acosta-Gonzalez G, Xu R. Clinical features, histology, and histogenesis of combined hepatocellular-cholangiocarcinoma. World J Hepatol. 2017; 9:300–309.
6. Akiba J, Nakashima O, Hattori S, Tanikawa K, Takenaka M, Nakayama M, et al. Clinicopathologic analysis of combined hepatocellular-cholangiocarcinoma according to the latest WHO classification. Am J Surg Pathol. 2013; 37:496–505.
7. Sasaki M, Sato H, Kakuda Y, Sato Y, Choi JH, Nakanuma Y. Clinicopathological significance of ‘subtypes with stem-cell feature’ in combined hepatocellular-cholangiocarcinoma. Liver Int. 2015; 35:1024–1035.
8. Sempoux C, Paradis V, Saxena R. Variant differentiation patterns in primary liver carcinoma. Semin Diagn Pathol. 2017; 34:176–182.
9. Komuta M, Spee B, Vander Borght S, De Vos R, Verslype C, Aerts R, et al. Clinicopathological study on cholangiolocellular carcinoma suggesting hepatic progenitor cell origin. Hepatology. 2008; 47:1544–1556.
10. Komuta M, Govaere O, Vandecaveye V, Akiba J, Van Steenbergen W, Verslype C, et al. Histological diversity in cholangiocellular carcinoma reflects the different cholangiocyte phenotypes. Hepatology. 2012; 55:1876–1888.
11. Moeini A, Sia D, Zhang Z, Camprecios G, Stueck A, Dong H, et al. Mixed hepatocellular cholangiocarcinoma tumors: cholangiolocellular carcinoma is a distinct molecular entity. J Hepatol. 2017; 66:952–961.
12. Maeno S, Kondo F, Sano K, Takada T, Asano T. Morphometric and immunohistochemical study of cholangiolocellular carcinoma: comparison with non-neoplastic cholangiole, interlobular duct and septal duct. J Hepatobiliary Pancreat Sci. 2012; 19:289–296.
13. Yeh MM. Pathology of combined hepatocellular-cholangiocarcinoma. J Gastroenterol Hepatol. 2010; 25:1485–1492.
14. Coulouarn C, Cavard C, Rubbia-Brandt L, Audebourg A, Dumont F, Jacques S, et al. Combined hepatocellular-cholangiocarcinomas exhibit progenitor features and activation of Wnt and TGFbeta signaling pathways. Carcinogenesis. 2012; 33:1791–1796.
15. Holczbauer Á, Factor VM, Andersen JB, Marquardt JU, Kleiner DE, Raggi C, et al. Modeling pathogenesis of primary liver cancer in lineage-specific mouse cell types. Gastroenterology. 2013; 145:221–231.
16. Li L, Qian M, Chen IH, Finkelstein D, Onar-Thomas A, Johnson M, et al. Acquisition of cholangiocarcinoma traits during advanced hepatocellular carcinoma development in mice. Am J Pathol. 2018; 188:656–671.
17. Zen C, Zen Y, Mitry RR, Corbeil D, Karbanová J, O’Grady J, et al. Mixed phenotype hepatocellular carcinoma after transarterial chemoembolization and liver transplantation. Liver Transpl. 2011; 17:943–954.
18. Fujii H, Zhu XG, Matsumoto T, Inagaki M, Tokusashi Y, Miyokawa N, et al. Genetic classification of combined hepatocellular-cholangiocarcinoma. Hum Pathol. 2000; 31:1011–1017.
19. Wang A, Wu L, Lin J, Han L, Bian J, Wu Y, et al. Whole-exome sequencing reveals the origin and evolution of hepato-cholangiocarcinoma. Nat Commun. 2018; 9:894.
20. Joseph NM, Tsokos CG, Umetsu SE, Shain AH, Kelley RK, Onodera C, et al. Genomic profiling of combined hepatocellular-cholangiocarcinoma reveals similar genetics to hepatocellular carcinoma. J Pathol. 2019; 248:164–178.
21. Cazals-Hatem D, Rebouissou S, Bioulac-Sage P, Bluteau O, Blanché H, Franco D, et al. Clinical and molecular analysis of combined hepatocellular-cholangiocarcinomas. J Hepatol. 2004; 41:292–298.
22. Fujimoto A, Furuta M, Shiraishi Y, Gotoh K, Kawakami Y, Arihiro K, et al. Whole-genome mutational landscape of liver cancers displaying biliary phenotype reveals hepatitis impact and molecular diversity. Nat Commun. 2015; 6:6120.
23. Liu ZH, Lian BF, Dong QZ, Sun H, Wei JW, Sheng YY, et al. Whole-exome mutational and transcriptional landscapes of combined hepatocellular cholangiocarcinoma and intrahepatic cholangiocarcinoma reveal molecular diversity. Biochim Biophys Acta Mol Basis Dis. 2018; 1864(6 Pt B):2360–2368.
24. Park SH, Lee SS, Yu E, Kang HJ, Park Y, Kim SY, et al. Combined hepatocellular-cholangiocarcinoma: gadoxetic acid-enhanced MRI findings correlated with pathologic features and prognosis. J Magn Reson Imaging. 2017; 46:267–280.
25. Kim JH, Yoon HK, Ko GY, Gwon DI, Jang CS, Song HY, et al. Non-resectable combined hepatocellular carcinoma and cholangiocarcinoma: analysis of the response and prognostic factors after transcatheter arterial chemoembolization. Radiology. 2010; 255:270–277.
26. Aoki K, Takayasu K, Kawano T, Muramatsu Y, Moriyama N, Wakao F, et al. Combined hepatocellular carcinoma and cholangiocarcinoma: clinical features and computed tomographic findings. Hepatology. 1993; 18:1090–1095.
27. Chantajitr S, Wilasrusmee C, Lertsitichai P, Phromsopha N. Combined hepatocellular and cholangiocarcinoma: clinical features and prognostic study in a Thai population. J Hepatobiliary Pancreat Surg. 2006; 13:537–542.
28. Ramai D, Ofosu A, Lai JK, Reddy M, Adler DG. Combined hepatocellular cholangiocarcinoma: a population-based retrospective study. Am J Gastroenterol. 2019; 114:1496–1501.
29. Yano Y, Yamamoto J, Kosuge T, Sakamoto Y, Yamasaki S, Shimada K, et al. Combined hepatocellular and cholangiocarcinoma: a clinicopathologic study of 26 resected cases. Jpn J Clin Oncol. 2003; 33:283–287.
30. Koh KC, Lee H, Choi MS, Lee JH, Paik SW, Yoo BC, et al. Clinicopathologic features and prognosis of combined hepatocellular cholangiocarcinoma. Am J Surg. 2005; 189:120–125.
31. Liu CL, Fan ST, Lo CM, Ng IO, Lam CM, Poon RT, et al. Hepatic resection for combined hepatocellular and cholangiocarcinoma. Arch Surg. 2003; 138:86–90.
32. Jarnagin WR, Weber S, Tickoo SK, Koea JB, Obiekwe S, Fong Y, et al. Combined hepatocellular and cholangiocarcinoma: demographic, clinical, and prognostic factors. Cancer. 2002; 94:2040–2046.
33. Palmer WC, Patel T. Are common factors involved in the pathogenesis of primary liver cancers? A meta-analysis of risk factors for intrahepatic cholangiocarcinoma. J Hepatol. 2012; 57:69–76.
34. Maeda T, Adachi E, Kajiyama K, Sugimachi K, Tsuneyoshi M. Combined hepatocellular and cholangiocarcinoma: proposed criteria according to cytokeratin expression and analysis of clinicopathologic features. Hum Pathol. 1995; 26:956–964.
35. Tang D, Nagano H, Nakamura M, Wada H, Marubashi S, Miyamoto A, et al. Clinical and pathological features of Allen’s type C classification of resected combined hepatocellular and cholangiocarcinoma: a comparative study with hepatocellular carcinoma and cholangiocellular carcinoma. J Gastrointest Surg. 2006; 10:987–998.
36. Lee WS, Lee KW, Heo JS, Kim SJ, Choi SH, Kim YI, et al. Comparison of combined hepatocellular and cholangiocarcinoma with hepatocellular carcinoma and intrahepatic cholangiocarcinoma. Surg Today. 2006; 36:892–897.
37. Kassahun WT, Hauss J. Management of combined hepatocellular and cholangiocarcinoma. Int J Clin Pract. 2008; 62:1271–1278.
38. Wells ML, Venkatesh SK, Chandan VS, Fidler JL, Fletcher JG, Johnson GB, et al. Biphenotypic hepatic tumors: imaging findings and review of literature. Abdom Imaging. 2015; 40:2293–2305.
39. Kim TH, Kim SY, Tang A, Lee JM. Comparison of international guidelines for noninvasive diagnosis of hepatocellular carcinoma: 2018 update. Clin Mol Hepatol. 2019; 25:245–263.
40. American College of Radiology. Liver imaging reporting and data system version 2018 [Internet]. Reston (USA): American College of Radiology;[cited 2020 Jun 30]. Available from: https://www.acr.org/Clinical-Resources/Reporting-and-Data-Systems/LI-RADS/CT-MRI-LI-RADS-v2018.
41. Marrero JA, Kulik LM, Sirlin CB, Zhu AX, Finn RS, Abecassis MM, et al. Diagnosis, staging, and management of hepatocellular carcinoma: 2018 practice guidance by the American Association for the Study of Liver Diseases. Hepatology. 2018; 68:723–750.
42. Fowler KJ, Sheybani A, Parker RA 3rd, Doherty S, Brunt EM, Chapman WC, et al. Combined hepatocellular and cholangiocarcinoma (biphenotypic) tumors: imaging features and diagnostic accuracy of contrast-enhanced CT and MRI. AJR Am J Roentgenol. 2013; 201:332–339.
43. de Campos RO, Semelka RC, Azevedo RM, Ramalho M, Heredia V, Armao DM, et al. Combined hepatocellular carcinoma-cholangiocarcinoma: report of MR appearance in eleven patients. J Magn Reson Imaging. 2012; 36:1139–1147.
44. Hwang J, Kim YK, Park MJ, Lee MH, Kim SH, Lee WJ, et al. Differentiating combined hepatocellular and cholangiocarcinoma from mass-forming intrahepatic cholangiocarcinoma using gadoxetic acid-enhanced MRI. J Magn Reson Imaging. 2012; 36:881–889.
45. Potretzke TA, Tan BR, Doyle MB, Brunt EM, Heiken JP, Fowler KJ. Imaging features of biphenotypic primary liver carcinoma (hepatocholangiocarcinoma) and the potential to mimic hepatocellular carcinoma: LI-RADS analysis of CT and MRI features in 61 cases. AJR Am J Roentgenol. 2016; 207:25–31.
46. Sammon J, Fischer S, Menezes R, Hosseini-Nik H, Lewis S, Taouli B, et al. MRI features of combined hepatocellular-cholangiocarcinoma versus mass forming intrahepatic cholangiocarcinoma. Cancer Imaging. 2018; 18:8.
47. Kim SS, Lee S, Choi JY, Lim JS, Park MS, Kim MJ. Diagnostic performance of the LR-M criteria and spectrum of LI-RADS imaging features among primary hepatic carcinomas. Abdom Radiol (NY). 2020; 45:3743–3754.
48. Jeon SK, Joo I, Lee DH, Lee SM, Kang HJ, Lee KB, et al. Combined hepatocellular cholangiocarcinoma: LI-RADS v2017 categorisation for differential diagnosis and prognostication on gadoxetic acid-enhanced MR imaging. Eur Radiol. 2019; 29:373–382.
49. Lee HS, Kim MJ, An C. How to utilize LR-M features of the LI-RADS to improve the diagnosis of combined hepatocellular-cholangiocarcinoma on gadoxetate-enhanced MRI? Eur Radiol. 2019; 29:2408–2416.
50. Mao Y, Xu S, Hu W, Huang J, Wang J, Zhang R, et al. Imaging features predict prognosis of patients with combined hepatocellular-cholangiocarcinoma. Clin Radiol. 2017; 72:129–135.
51. An C, Park S, Chung YE, Kim DY, Kim SS, Kim MJ, et al. Curative resection of single primary hepatic malignancy: liver imaging reporting and data system category LR-M portends a worse prognosis. AJR Am J Roentgenol. 2017; 209:576–583.
52. Gigante E, Ronot M, Bertin C, Ciolina M, Bouattour M, Dondero F, et al. Combining imaging and tumour biopsy improves the diagnosis of combined hepatocellular-cholangiocarcinoma. Liver Int. 2019; 39:2386–2396.
53. Nahm JH, Rhee H, Kim H, Yoo JE, Lee JS, Jeon Y, et al. Increased expression of stemness markers and altered tumor stroma in hepatocellular carcinoma under TACE-induced hypoxia: a biopsy and resection matched study. Oncotarget. 2017; 8:99359–99371.
Figure 1
A 57-year-old male with combined hepatocellular-cholangiocarcinoma (cHCC-CCA) in the background liver of hepatitis B-virus and alcohol related chronic hepatitis. A 3.4-cm infiltrative mass lesion in the liver segment 7 shows low-signal intensity in precontrast T1-weighted image (A), peripheral enhancement in the arterial phase (B), absence of washout in portal phase (C) and 2-minute delay phase (D), decreased hepatobiliary uptake in hepatobiliary phase (E), and high-signal intensity in T2-weighted image (F) of gadoxetate-enhanced magnetic resonance imaging. It is categorized as Liver Imaging Reporting and Data System M based on targetoid appearance. On pathologic examination (G–I), the tumor shows cholangiocarcinoma (CCA) component in whitish and fibrotic area of gross specimen and hepatocellular carcinoma (HCC) component in more yellowish area of resected specimen, and there are transitional differentiation zones between them (G, gross feature of resected specimen; H, scanning view of hematoxylin-eosin stain; I, map of histological components). HCC and CCA areas are not distinguishable in magnetic resonance imaging.
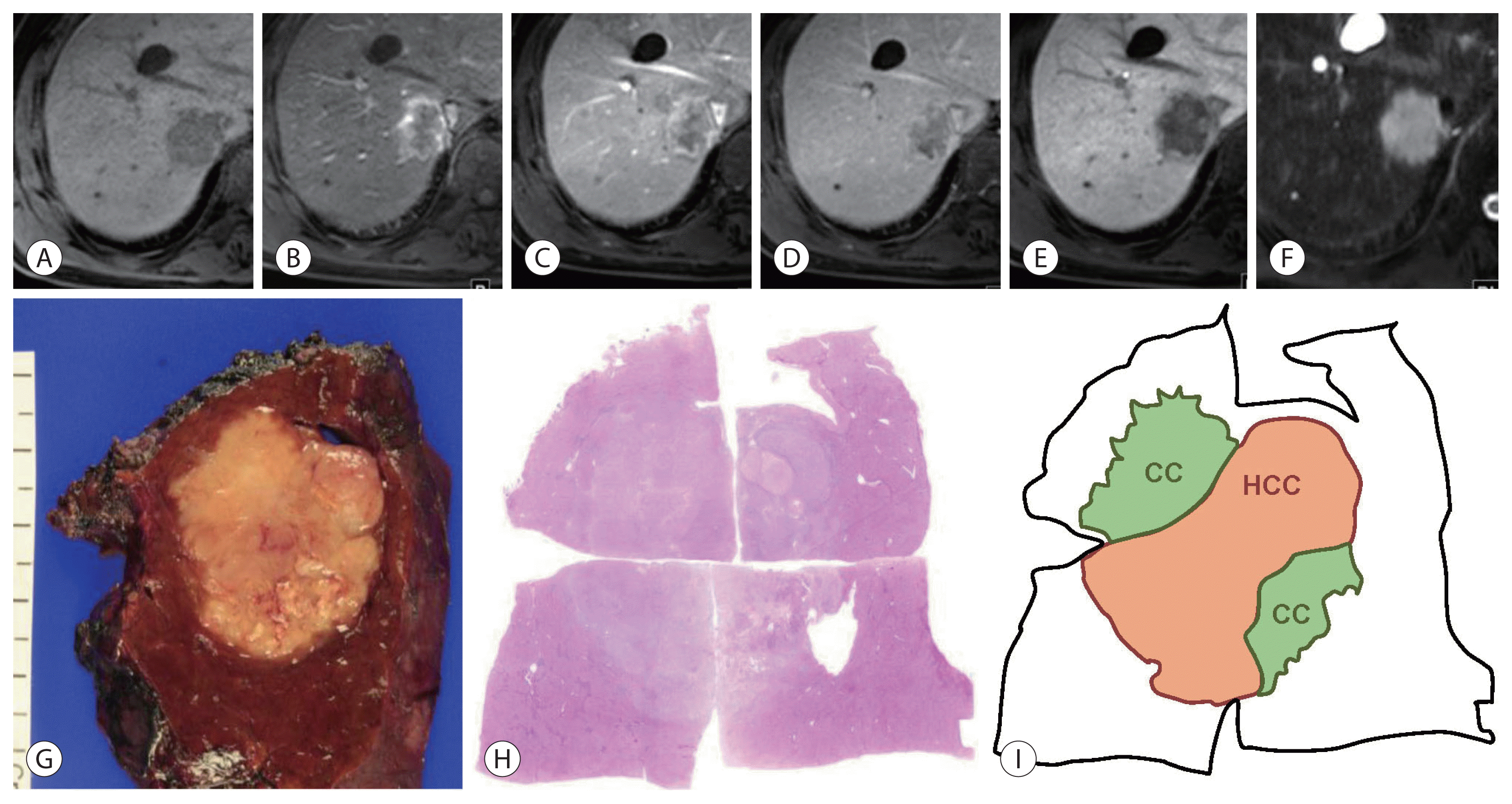
Figure 2
A 58-year-old male showing combined hepatocellular-cholangiocarcinoma (cHCC-CCA) with cholangiolocellular (CLC) and intermediate-cell carcinoma components, developed in cirrhosis of unknown etiology. A 3.2-cm lobulated mass lesion in the liver segment 4 shows low-signal intensity in precontrast T1-weighted image (A), peripheral enhancement in the arterial phase (B), absence of washout in portal phase (C) and delayed central enhancement in hepatobiliary phase (D) of gadoxetate-enhanced magnetic resonance imaging (MRI). The lesion also shows nodule-in-nodule appearance: the inner nodule exhibited hypervascularity in arterial phase, absence of washout in portal phase, and low-signal intensity in T2-weighted image (E). As the lesion shows peripheral arterial enhancement and delayed central enhancement, it is categorized as Liver Imaging Reporting and Data System M. On pathologic examination (F–L), the tumor shows complex mixture of hepatocellular carcinoma (HCC) (I), intermediate-cell carcinoma (J), CLC (K), and cholangiocarcinoma (CCA) (L) components. On MRI, the CCA component, corresponding the inner nodule, was more hypervascular than other components including HCC ([F] gross feature of resected specimen; [G] scanning view of hematoxylin-eosin [H–E] stain; [H] map of histological components; [I–L] H–E stain, original magnification, ×100).
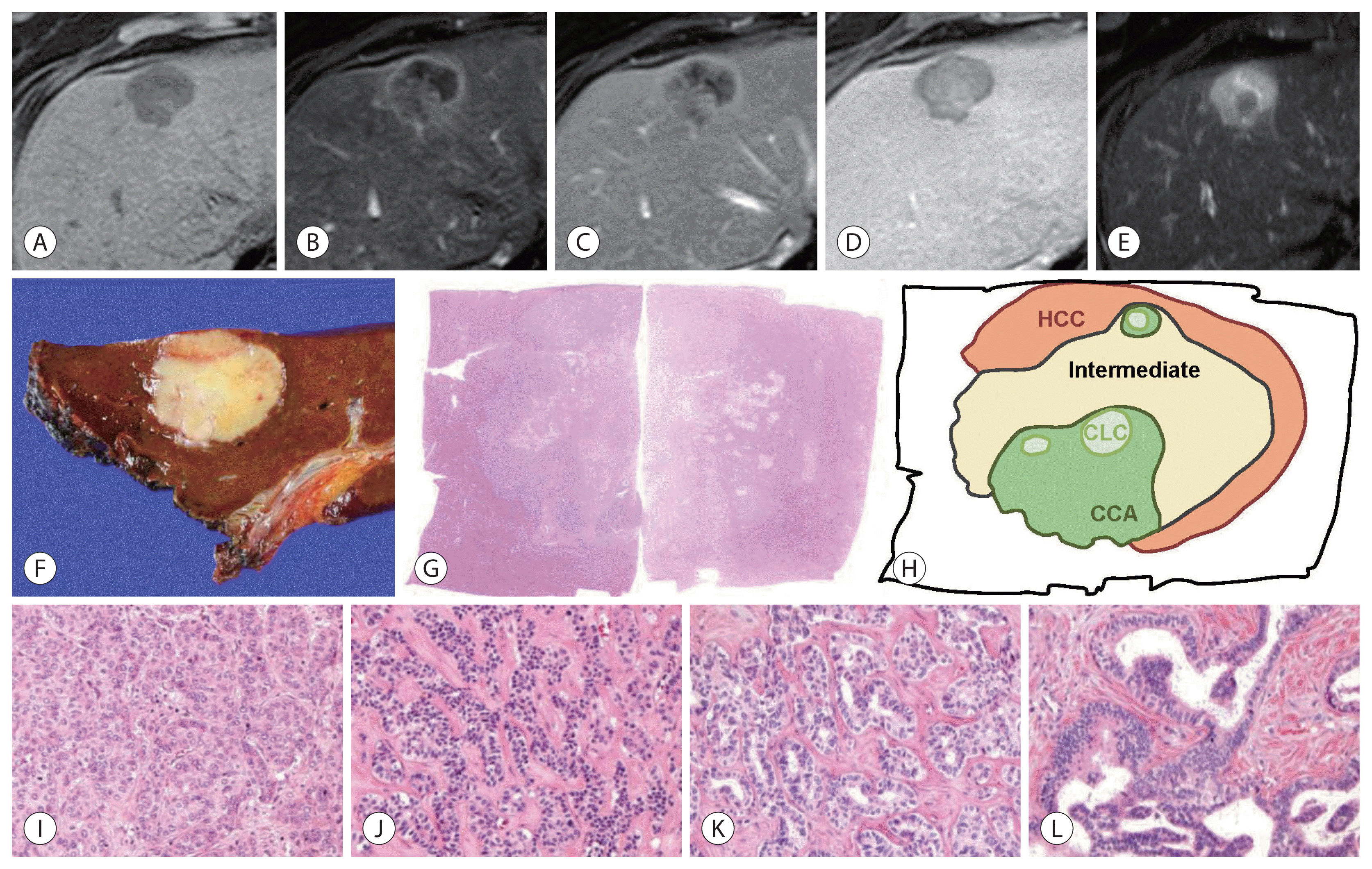
Figure 3
A 50-year-old female showing combined hepatocellular-cholangiocarcinoma (cHCC-CCA), developed in B viral cirrhosis. A 1.7-cm tumor in the liver segment 2 shows low-signal intensity on precontrast T1-weighted image (A), non-peripheral enhancement in the arterial phase (B), washout and enhancing capsule on portal phase (C) and 2-minutes delay phase (D) of gadoterate meglumine-enhanced magnetic resonance imaging (white arrows). The lesion also shows high signal intensity in T2-weighted image (E) and diffusion-weighted image (F, b=800). Since the lesion exhibits three major features of Liver Imaging and Reporting and Data System, it is categorized as HCC (LR-5). On pathologic examination, the tumor is composed of two histologic components (G–L), showing large area of HCC (J) and small area of CCA (K) on hematoxylin-eosin (H–E) staining. Alcian-blue staining shows mucin, stained as blue color, at CCA (L, red arrows) ([G] gross feature of resected specimen; [H] scanning view of H–E stain; [I] map of histological component; [J, K] H–E stain, original magnification, ×100; [L] scanning view of Alcian-blue stain).
Figure 4
A 67-year-old male with combined hepatocellular-cholangiocarcinoma (cHCC-CCA), developed in C viral cirrhosis with matched biopsied and resected specimen. The gadoxetate-enhanced magnetic resonance imaging shows approximately 2.4 cm mass lesion with low-signal intensity in precontrast T1-weighted image (A), peripheral enhancement in the arterial phase (B), absence of washout in portal phase (C) and decreased hepatobiliary uptake in hepatobiliary phase (D), high signal intensity in T2-weighted image (E), and targetoid restriction in diffusion-weighted image (F, b=800). The patient has past history of distal common bile duct cancer (26 years ago), and recently diagnosed squamous cell carcinoma of oral cavity. To determine whether the lesion is metastasis, percutaneous liver biopsy was performed. The biopsy specimen shows adenocarcinoma without other component (G, H). The patient underwent hepatic resection and the tumor reveals cHCC-CCA (I, J). There is large area of cholangiocarcinoma (approximately 90%) and small area of hepatocellular carcinoma (approximately 10%) with transitional differentiation zones between them ([G] hematoxylin-eosin stain [H–E], original magnification, ×100; [H, I] scanning view of H–E stain; [J] H–E stain, original magnification, ×40).

Table 1
Summary of published molecular features of cHCC-CCA
| Reference | Technique | Number of cHCC-CCA | Molecular features |
|---|---|---|---|
| Fujii et al.18 (2000) | Loss of heterozygosity | 8 | HCC and CCA in cHCC-CCA shared allelic losses, suggesting clonality. |
| Cazals-Hatem et al.21 (2004) | Loss of heterozygosity, Sanger sequencing | 15 | Mutation patterns, allelic losses were closer to CCA. |
| Coulouarn et al.14 (2012) | Gene expression array | 20 |
Stem/progenitor feature, down-regulation of hepatocyte differentiation related genes, up-regulation biliary differentiation related genes. TGF-β and Wnt/β-catenin signaling activation were the major pathways of cHCC-CCA. |
| Fujimoto et al.22 (2015) | Whole genome sequencing | 7 | Mutation patterns and allelic losses of cHCC-CCA with chronic hepatitis were close to HCC. Those without chronic hepatitis were diverse. |
| Moeini et al.11 (2017) | Gene expression array, Whole-exome sequencing, Genome-wide analysis | 18 |
CLC showed enriched TGF-β signaling and biliary-like feature compared to other types of cHCC-CCAs. cHCC-CCA with stem cell features often showed SALL4 expression, progenitor-like signature and poor prognostic signature. Classical type tumors showed biphenotypic profile, and shared copy number variants, suggesting clonality. |
| Sasaki et al.7 (2015) | Sanger sequencing | 53 | cHCC-CCA exhibited diverse mutations, which might reflect the etiological and histological subtypes, and tumor aggressiveness. |
| Wang et al.19 (2018) | Whole-exome sequencing | 15 |
HCC and CCA in cHCC-CCA showed both of synonymous and non-synonymous genetic alterations, suggesting clonality and intra-tumoral genetic heterogeneity. cHCC-CCA showed expression of stem/progenitor markers. |
| Liu et al.23 (2018) | Whole-exome sequencing, RNA sequencing | 10 | Mutation and transcription patterns were closer to HCC. |
| Joseph et al.20 (2019) | Capture-based DNA sequencing | 20 | Mutation patterns were close to HCC. |
Table 2
Summary of published radiologic features of cHCC-CCA
| Reference | Technique | Number of cHCC-CCA | Radiologic features | LR-M features | Other features |
|---|---|---|---|---|---|
| Major features | |||||
| Fowler et al.42 (2013) | CT or MRI | 29 | |||
| de Campos et al.43 (2012) | MRI | 11 | |||
| Hwang et al.44 (2012) | MRI | 20 | |||
| Wells et al.38 (2015) | CT or MRI | 29 | |||
| Potretzke et al.45 (2016) | CT or MRI | 61 | |||
| Park et al.24 (2017) | MRI | 82 | |||
| Sammon et al.46 (2018) | MRI | 33 | |||
| Jeon et al.48 (2019) | MRI | 70 | |||
| Kim et al.47 (2020) | MRI | 43 |
Table 3
Comparison of clinical, histological, and radiological characters of primary liver cancers




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download