Abstract
Objectives
The primary aim of this study is to evaluate the gene expression profile of Asian sand dust (ASD)-treated human middle ear epithelial cell (HMEEC) using microarray analysis.
Methods
The HMEEC was treated with ASD (400 µg/mL) and total RNA was extracted for microarray analysis. Molecular pathways among differentially expressed genes were further analyzed. For selected genes, the changes in gene expression were confirmed by real-time polymerase chain reaction.
Results
A total of 1,274 genes were differentially expressed by ASD. Among them, 1,138 genes were 2 folds up-regulated, whereas 136 genes were 2 folds down-regulated. Up-regulated genes were mainly involved in cellular processes, including apoptosis, cell differentiation, and cell proliferation. Down-regulated genes affected cellular processes, including apoptosis, cell cycle, cell differentiation, and cell proliferation. The 10 genes including ADM, CCL5, EDN1, EGR1, FOS, GHRL, JUN, SOCS3, TNF, and TNFSF10 were identified as main modulators in up-regulated genes. A total of 11 genes including CSF3, DKK1, FOSL1, FST, TERT, MMP13, PTHLH, SPRY2, TGFBR2, THBS1, and TIMP1 acted as main components of pathway associated with 2-fold down regulated genes.
Sand storms arise very common in desert area and in Asia, it occurs in Gobi Desert, the Taklimakan Desert and interior China. Asian sand dust (ASD) spreads neighboring area including East China, Korean Peninsula, and Japan [1]. ASD is composed of various chemical components such as Sulfate (SO42) and Nitrate (NO3-) derived from air pollutants and microbial agents, including bacteria, fungi, fungal spores, and viruses [2]. ASD is known to be related with enhancement of ovalbumin induced lung eosinophilia, chronic pulmonary injury [3], and rhinovirus replication in human nasal epithelial cells [4]. Therefore, hazardous effects of ASD on human health are becoming a serious concern in East Asia.
The otitis media (OM) is multifactorial disease and the causative agent includes a viral and bacterial infection, biofilm formation, congenital anomaly, and environmental factor, such as smoking, and air pollution [5].
The epidemiologic studies have investigated the relationship between ambient air pollution and OM in human populations. A study conducted in the Czech Republic showed that children living in areas with high concentrations of particulate matter (PM) and SO2 have significantly higher rates of OM compared with those in the control area [6]. Rover et al. [7] showed that the level of SO2 emission is one of the important risk factors of OM in many industrialized western countries. A large cohort study showed the close relationship between the prevalence of OM and air quality [8].
In our previous studies, we have shown that air pollutant such as urban particle, diesel exhaust particle and acrolein can induced inflammation and cause alteration of gene expression human middle ear epithelial cell (HMEEC) [910]. However, there is no study about the biological effect of ASD on middle ear epithelial cells.
The aim of this study is to evaluate the gene expression profile of ASD-treated HMEEC using microarray analysis.
ASD were collected on March 16, 2009 at outside the Gachon University building in Korea using a high volume air sampler (HV500F, Sibata, Tokyo, Japan) at a flow rate of 500 L/minute. These ASD were prefiltered to filter packs (prefilter AP, 124 mm; Millipore, Bedford, MA, USA) and then sieved on filter of 10-µm pore size after mixing with phosphate buffered saline in a 10-mL tube. The filtered ASD particles were sterilized in an autoclave at 121℃ for 15 minutes and determined their mass and then collected in a 1.5-mL tube. The filtered PM was stored at -20℃ until needed. The particle diameter of the samples (a total of 600 particles) was measured under a scanning electron microscope (JSM-5800 JEOL Ltd., Tokyo, Japan). The mean distribution peak of particle diameter in ASD was observed at 6 mm (Fig. 1). Chemical component of ASD particles were analyzed with X-ray fluorescence spectrometry (PHILIPS PW2404, PHILIPS Electronic Instruments, Amsterdam, the Netherlands) at Korea basic science institute.
HMEECs were maintained in a mixture of high glucose Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA, USA) and bronchial epithelial basal medium (Lonza, Walkersville, MD, USA) (1:1). The cells were maintained at subconfluence in a humidified atmosphere of 5% CO2 and 95% air at 37°C.
The cells were grown to 60% confluence in six-well culture plates. After starvation for 2 hours, medium was replaced to ASD containing medium and exposed to disperse ASD at a final concentration of 400 µg/mL for 24 hours. For the control group, ASD was not added.
Total RNAs were extracted using Trizol reagent (Ambion, Carlsbab, CA, USA), and cDNA was synthesized by AccuPower reverse transcriptase premix (BIONEER, Daejeon, Korea). Three RNA samples were used for miRNA microarray analysis. The RNA samples were then labeled with Cyanine 3-CTP (Cy3) and Cyanine 5-CTP (Cy5) in an in vitro transcription reaction using a Low Input Quick Amp Labeling kit (Agilent Technologies, Santa Clara, CA, USA), in accordance with the manufacturer's protocol. Labeled cRNA was hybridized on Human GE 4×44K v2 microarray (ID G2519-026652, Agilent Technologies), followed by manual washing, according to the manufacturer's procedures. The array was scanned using the Agilent DNA MicroArray Scanner and probe signals were quantified using Agilent's Feature Extraction ver. 10.10.1.1 (Agilent Technologies). Normalized data were analyzed using Subio platform v1.16.4376 (Subio Inc., Kagoshima, Japan).
Molecular pathways, among the differentially expressed genes identified by the microarray, were dissected using Pathway Studio 9.0 (Ariadne Genomics, Rockville, MD, USA). This program used to facilitate integration of relevant information among the imported genes, consequently allowing identification of biological pathways, gene regulation networks and protein interaction maps.
Total RNA was extracted using Trizol according to the protocol of the manufacturer. Yield and purity was determined by spectro-photometry and total RNA was reverse transcribed to cDNA using PrimeScript 1st Strand cDNA Synthesis kit according to the manufacturer's instructions (TaKaRa Bio Inc., Kusatsu, Japan). The quantitation of mRNA expression was carried out using the ABI Prism 7300 real-time polymerase chain reaction (PCR) system (Applied Biosystems, Foster, CA, USA). The PCR amplification performed with a 20-µL final reaction mixture and PCR reaction mixture were incubated at 95℃ for 15 seconds and 60℃ for 1 minutes, followed by amplification for 45 cycles. The relative level of gene expressions were normalized to endogenous glyceraldehyde-3-phosphate dehydrogenase and target mRNA expression in the experimental groups were calculated, relative to the control group.
The genes and primers used for RT-PCR were as follows: MUC1 (mucin 1, cell surface associated), MUC4 (mucin 4, cell surface associated), TNF (tumor necrosis factor [TNF superfamily, member 2]), TNFSF14 (tumor necrosis factor [ligand] superfamily, member 14), CCL5 (chemokine [C-C motif] ligand 5), CCL20 (chemokine [C-C motif] ligand 20), CXCL10 (chemokine [C-X-C motif] ligand 10), JUN (jun oncogene), ANGPT1 (angiopoietin 1), and TIMP1 (TIMP metallopeptidase inhibitor 1) (Table 1).
The most abundant component was 78.4% for SiO2 among the contents of elements in ASD. Other ASD components were 9.35% for Al2O3, 2.52% for K2O, 2.41% for Na2O, 2.06% for Fe2O3, 1.74% for CaO, respectively. MaO, TiO2, P2O5, and MnO composed less than 0.1% of the total (Table 2).
We performed microarray analysis and depicted molecular pathway using Pathway Studio 9.0 to characterize the effects of ASD exposure to HMEEC. Microarray data assessed the number of genes that showed a fold change of >2 or <0.5 in each group compared with the control group. A total of 1,274 genes were differentially regulated after exposure of ASD. Among them, 1,138 genes were significantly up-regulated, whereas 136 genes were down-regulated. ASD exposure showed a prominent increase in the gene expression involved in signal transduction and regulation of metabolism pathway.
A total of 1,274 genes were classified in up-regulated genes and down-regulated genes, it represents biological network of genes in HMEECs exposed to ASD. Up-regulated genes were mainly involved in cellular processes, including cell proliferation, apoptosis, cell differentiation, cell growth, cell death (Fig. 2). Down-regulated genes affected several cellular processes, including cell cycle, cell proliferation, apoptosis and cell growth (Fig. 3). Gene ontology analysis of microarray data also showed the biological functions of the differentially expressed gene signature in HMEECs treated ASD (Fig. 4).
We investigated signaling networks using Pathway Studio 9.0 to determine potential molecular signaling networks by ASD exposure. Pathway Studio 9.0 represented that relevant components in the putative signaling pathways were chosen and incorporated into the established networks, based on a number of reliable studies.
A total of 47 genes were identified as crucial components in our signaling networks data containing 2-fold up regulated genes (Table 3, Fig. 2). Among them, 10 genes including ADM, CCL5, EDN1, EGR1, FOS, GHRL, JUN, SOCS3, TNF, and TNFSF10 were discovered as the key mediator genes among the up-regulated genes altered by ASD exposure. These 10 genes are shown to be related to diverse biological processes, including cell proliferation, cell differentiation, apoptosis, cell growth and cell death and these cell processes might lead to neoplasm, inflammation, infection, cancer, and death (Fig. 2).
On the other hand, a total of 38 genes were revealed as key modulators in the signaling pathway associated with 2-fold down regulated genes (Table 4, Fig. 3). Among 38 down-regulated genes, 11 genes, CSF3, DKK1, FOSL1, FST, TERT, MMP13, PTHLH, SPRY2, TGFBR2, THBS1, and TIMP1, were identified as the main modulator genes. These down-regulated gene sets showed distinct molecular pathway related to cell differentiation, cell cycle, apoptosis, cell proliferation, cell growth, which can induce neoplasm, neoplasm metastasis, inflammation, cancer, death, wounds and injuries (Fig. 3).
Recently, the environmental factor, such as air pollution and PM have been evaluated for hazardous effects on human health and most of these studies are elucidating the correlation between environmental factors and human disease [1]. In the present study, we focused on the effects of ASD on HMEECs to determine the relationship between ASD and the pathogenesis of OM in human.
ASD are mainly composed of various chemical components as well as biological materials such as bacterial, fungi and viruses. The composition of ASD mix can change during their long-range transport and it can cause numerous harmful health effect according to materials of ASD mix [11]. Recent studies reported that ASD exposure is associated with inflammatory reaction via cytokine production [12]. Also, it has been reported that acute and chronic pulmonary toxicity was induced by intratracheally instilled ASD [3]. However, specific molecular signaling network for ASD-induced toxic response in human middle ear epithelium is not investigated yet. We, therefore, analyzed gene expression profile using microarray to understand molecular mechanism between ASD and OM. Visualization of signaling pathways among differentially expressed genes enables to create their own pathways and understand complicated signaling network easily [13]. Additionally, Pathway Studio 9.0 was applied for investigation of molecular signaling network triggered by ASD in human cells.
A total of 1,274 genes were 2 folds differentially expressed by ASD exposure. Among them, 1,138 genes were 2 folds up-regulated, whereas 136 genes were 2 folds down-regulated. Our signaling network among 2-fold up-regulated genes suggested several cellular processes including apoptosis, cell death, cell differentiation, cell growth, and cell proliferation might be regulated by ASD exposure. Also, this cell processes may lead to cancer, death, inflammation, infection, and neoplasm. Our signaling network data suggested the key modulators based on integration of relevant information among imported genes, consequently al-lowing identification of biological pathways, gene regulation networks, and protein interaction maps. The 10 genes including ADM, CCL5, EDN1, EGR1, FOS, GHRL, JUN, SOCS3, TNF, and TNFSF10 were identified as main modulators in up-regulated genes. TNF family genes are a multifunctional proinflammatory cytokine. Regarding PM, few of studies have been reported recently. TNF-α production was enhanced by yellow sand dust exposure in RBL-2H3 cells [14]. Another study reported increase of TNF-α in bronchoalveolar lavage under ASD exposure [15]. Our data also showed up-regulation of TNF genes under ASD exposure, therefore, this gene can be a critical biomarker for ASD.
Yanagisawa et al. [16], examined the enhancement of inflammatory response-related genes in the murine lung by ASD using microarray analysis. Inflammation is a critical for OM, and uncontrolled inflammation in the middle ear can cause the development and progression of OM. In our data, inflammatory response-related genes including CCL5, CCL20, and CXCL10 are increased after exposure to ASD to HMEEC compared to the control group, indicating that ASD-induced inflammatory cytokines can affect to the development of OM. One of the key modulators in up-regulated genes, JUN gene, is known to be regulated by peptide growth factors, proinflammatory cytokines, oxidative and other forms of cellular stress with dimerization of AP-1 [17]. In our data, JUN was up-regulated by ASD, indicating that it may act as a main mediator to induce OM via regulation of inflammatory cytokines and oxidative stress in middle ear epithelial cells.
Mucins are known to play an important role for the protection and function of the underlying middle ear epithelium. In this study, MUC1 and MUC4 expression level were higher after ASD exposure. It can relate to mucous production for protection of epithelium by ASD. Wei et al. [18], reported interaction of MUC1 with p53 results in inhibition of p53-mediated apoptosis and cell cycle arrest. In our data, MUC1 was increased in HMEECs exposed ASD compared to the control group. It means that MUC1 may contribute to a defense mechanism to prevent cell death against ASD exposure.
Other genes, ADM, CCL5, EDN1, EGR1, FOS, GHRL, SOCS3, and TNFSF10, were firstly identified and suggested as responsible genes toward ASD exposure. Therefore, further studies are warranted for being reliable biomarker.
We also depicted molecular signaling network among 2-fold down-regulated genes. We found that 11 genes including CSF3, DKK1, FOSL1, FST, TERT, MMP13, PTHLH, SPRY2, TGFBR2, THBS1, and TIMP1 acted as main components of pathway as-sociated with apoptosis, cell cycle, cell growth, cell differentiation, and cell proliferation against ASD exposure. Among them, FOSL1 was significantly up-regulated against PM 2.5 in A549 cells [19]. Although our data showed down-regulation of FOSL1, this gene might be considered as a candidate ASD biomarker.
Telomerase reverse transcriptase (TERT) has been known to prevent of aging following multiple round of replication. Recent study demonstrated that TERT partially protect HSCs from reactive oxygen species-induced apoptosis via p38MAPK activation [20]. Expression of TERT was decreased compared to the control group in our present study. Therefore, down-regulation of TERT might affect reactive oxygen species-induced apoptosis and block cell survival in HMEECs after ASD exposure. In addition down-regulation of TIMP1 has been suggested as key modulator toward micro particle in HMEECs [10]. Similarly, our result showed down-regulated TIMP1 against ADS exposure. Hence, this gene can be candidates for potential biomarker. Newly identified genes, CSF3, DKK1, FST, TERT, MMP13, PTHLH, SPRY2, TGFBR2, and THBS1, might be suggested as significant modulators against ADS.
To compare with previous our study about micro particle in HMEECs, both PM and ASD showed similar gene expression profile in several cellular processes including apoptosis, cell differentiation, cell proliferation, and cell growth. However, ASD exposure markedly enhanced mucin genes and inflammatory response-related genes based on the 24-hour data. These results indicate that common and unique genes and signaling pathways affect biological effect on HMEECs according to different particle compositions between PM and ASD.
ACKNOWLEDGMENTS
This study was supported by the Korea Ministry of Environment as part of "The Environmental Health Action Program" (2012 001360002).
References
1. He M, Ichinose T, Yoshida S, Yamamoto S, Inoue K, Takano H, et al. Asian sand dust enhances murine lung inflammation caused by Klebsiella pneumoniae. Toxicol Appl Pharmacol. 2012; 1. 258(2):237–247. PMID: 22118940.

2. Chen PS, Tsai FT, Lin CK, Yang CY, Chan CC, Young CY, et al. Ambient influenza and avian influenza virus during dust storm days and background days. Environ Health Perspect. 2010; 9. 118(9):1211–1216. PMID: 20435545.

3. Naota M, Shiotsu S, Shimada A, Kohara Y, Morita T, Inoue K, et al. Pathological study of chronic pulmonary toxicity induced by intratracheally instilled Asian sand dust (kosa). Toxicol Pathol. 2013; 1. 41(1):48–62. PMID: 22744225.

4. Yeo NK, Hwang YJ, Kim ST, Kwon HJ, Jang YJ. Asian sand dust enhances rhinovirus-induced cytokine secretion and viral replication in human nasal epithelial cells. Inhal Toxicol. 2010; 10. 22(12):1038–1045. PMID: 20879958.

5. Yadav MK, Chae SW, Song JJ. In vitro Streptococcus pneumoniae biofilm formation and in vivo middle ear mucosal biofilm in a rat model of acute otitis induced by S. pneumoniae. Clin Exp Otorhinolaryngol. 2012; 9. 5(3):139–144. PMID: 22977710.
6. Dostal M, Hertz-Picciotto I, James R, Keller J, Dejmek J, Selevan S, et al. Childhood morbidity and air pollution in the Teplice program. Cas Lek Cesk. 2001; 10. 140(21):658–661. PMID: 11766454.
7. Rovers MM, de Kok IM, Schilder AG. Risk factors for otitis media: an international perspective. Int J Pediatr Otorhinolaryngol. 2006; 7. 70(7):1251–1256. PMID: 16481051.

8. Heinrich J, Frye C, Holscher B, Meyer I, Pitz M, Cyrys J, et al. Environmental surveys in the areas of Bitterfeld, Hettstedt and a comparative area in 1992-2000. Gesundheitswesen. 2002; 12. 64(12):675–682. PMID: 12516020.
9. Song JJ, Lee JD, Lee BD, Chae SW, Park MK. Effect of acrolein, a hazardous air pollutant in smoke, on human middle ear epithelial cells. Int J Pediatr Otorhinolaryngol. 2013; 10. 77(10):1659–1664. PMID: 23953484.

10. Song JJ, Kwon JY, Park MK, Seo YR. Microarray analysis of gene expression alteration in human middle ear epithelial cells induced by micro particle. Int J Pediatr Otorhinolaryngol. 2013; 10. 77(10):1760–1764. PMID: 24012219.

11. Ichinose T, Nishikawa M, Takano H, Sera N, Sadakane K, Mori I, et al. Pulmonary toxicity induced by intratracheal instillation of Asian yellow dust (Kosa) in mice. Environ Toxicol Pharmacol. 2005; 7. 20(1):48–56. PMID: 21783567.

12. Kang IG, Jung JH, Kim ST. Asian sand dust enhances allergen-induced th2 allergic inflammatory changes and mucin production in BALB/c mouse lungs. Allergy Asthma Immunol Res. 2012; 7. 4(4):206–213. PMID: 22754714.

13. Nikitin A, Egorov S, Daraselia N, Mazo I. Pathway studio: the analysis and navigation of molecular networks. Bioinformatics. 2003; 11. 19(16):2155–2157. PMID: 14594725.
14. Yamada P, Hatta T, Du M, Wakimizu K, Han J, Maki T, et al. Inflammatory and degranulation effect of yellow sand on RBL-2H3 cells in relation to chemical and biological constituents. Ecotoxicol Environ Saf. 2012; 10. 84:9–17. PMID: 22835726.

15. Song Y, Ichinose T, Morita K, Nakanishi T, Kanazawa T, Yoshida Y. Asian sand dust causes subacute peripheral immune modification with NF-κB activation. Environ Toxicol. 2015; 5. 30(5):549–558. PMID: 24376072.

16. Yanagisawa R, Takano H, Ichinose T, Mizushima K, Nishikawa M, Mori I, et al. Gene expression analysis of murine lungs following pulmonary exposure to Asian sand dust particles. Exp Biol Med (Maywood). 2007; 9. 232(8):1109–1118. PMID: 17720957.

17. Wisdom R, Johnson RS, Moore C. c-Jun regulates cell cycle progression and apoptosis by distinct mechanisms. EMBO J. 1999; 1. 18(1):188–197. PMID: 9878062.

18. Wei X, Xu H, Kufe D. Human MUC1 oncoprotein regulates p53-responsive gene transcription in the genotoxic stress response. Cancer Cell. 2005; 2. 7(2):167–178. PMID: 15710329.

19. Gualtieri M, Longhin E, Mattioli M, Mantecca P, Tinaglia V, Mangano E, et al. Gene expression profiling of A549 cells exposed to Milan PM2.5. Toxicol Lett. 2012; 3. 209(2):136–145. PMID: 22178795.

20. Nitta E, Yamashita M, Hosokawa K, Xian M, Takubo K, Arai F, et al. Telomerase reverse transcriptase protects ATM-deficient hematopoietic stem cells from ROS-induced apoptosis through a telomere-independent mechanism. Blood. 2011; 4. 117(16):4169–4180. PMID: 21297001.

Fig. 1
Analysis of Asian sand dust (ASD) constitution. (A) Scanning electron microscopic photomicrograph of ASD particles. (B) Distribution peak of particle diameter in ASD.
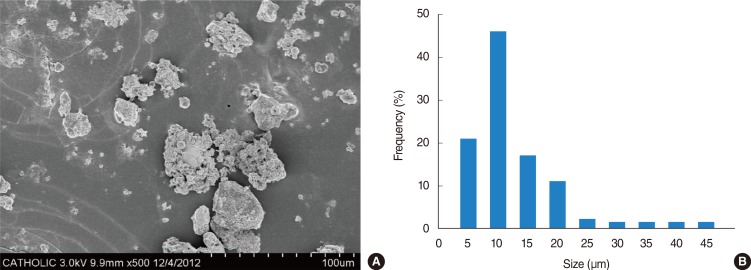
Fig. 2
Dissection of responsive molecular network of up-regulated genes induced by Asian sand dust. Magnified red circles indicate up-regulated genes in our microarray data; EGR1, CCL5, TNF, FOS, ADM, EDN1, TNFSF10, SOCS3, CCL11, MAP3K8, JUN, TGFB1, MAPK8, MAPK14, and GHRL.
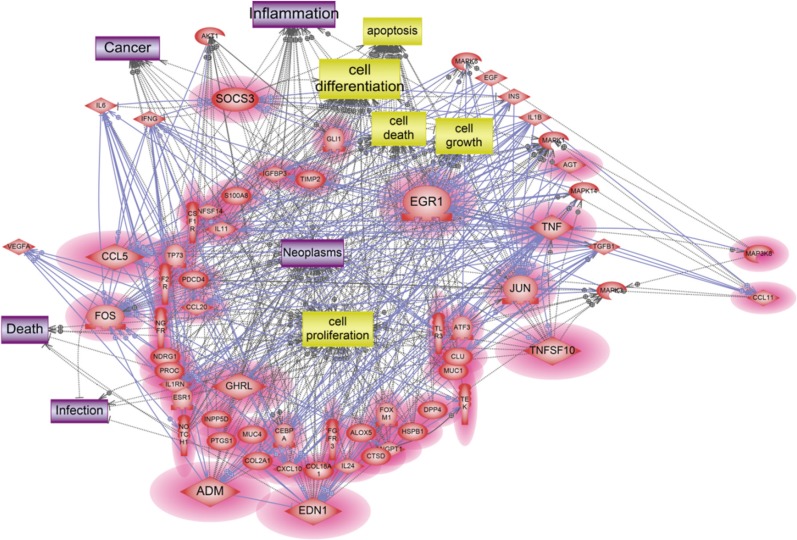
Fig. 3
Dissection of responsive molecular network of down-regulated genes induced by Asian sand dust. Magnified blue circles indicate down-regulated genes in our microarray data; FST, TERT, SPRY2, PTHLH, MMP13, DKK1, TGFBR2, CSF3, FOSL1, THBS1, MAPK1, MAPK1, SH202A, and TIMP1.
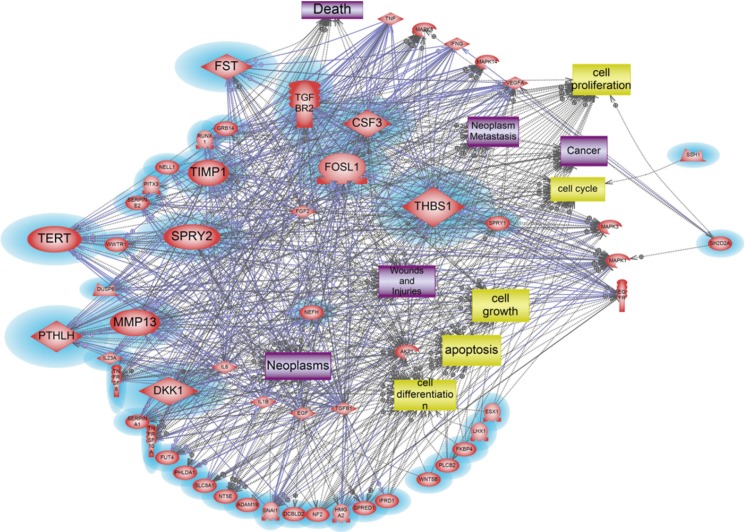
Fig. 4
Biological processes involving genes that are altered in response to Asian sand dust exposure at human middle ear epithelial cells are shown in graph.
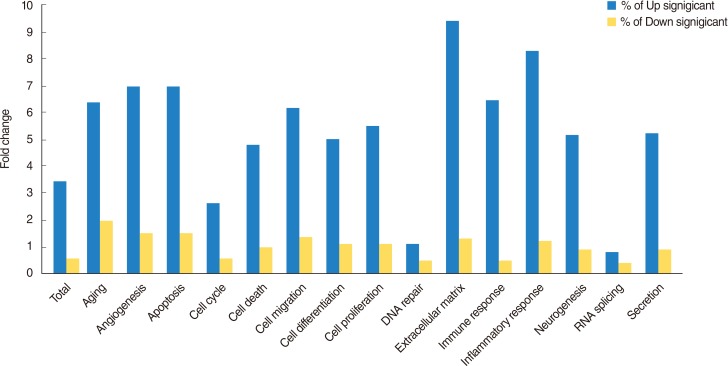
Fig. 5
Quantitative reverse transcription polymerase chain reaction analysis of selected genes at 24 hours after exposure to Asian sand dust (ASD). The results are shown as means±SD. All data are determined P-value. P<0.05 vs. the control group.
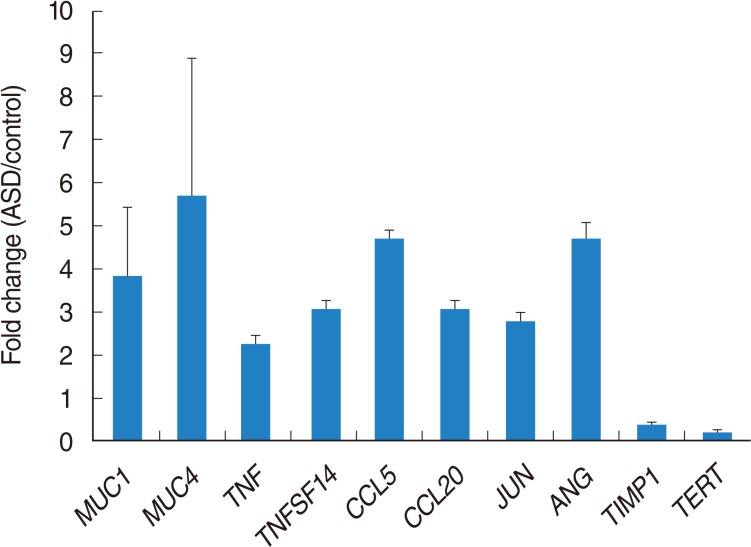
Table 1.
Sequences of primers used for real-time reverse transcription polymerase chain reaction
Table 2.
Elemental contents of Asian sand dust
| Component | Element fraction (wt%) |
|---|---|
| SiO2 | 78.4 |
| Al2O3 | 9.35 |
| K2O | 2.52 |
| Na2O | 2.41 |
| Fe2O3* | 2.06 |
| CaO | 1.74 |
| MgO | 0.69 |
| TiO2 | 0.29 |
| P2O5 | 0.05 |
| MnO | 0.05 |
| Loss on ignition | 1.74 |




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download