Abstract
Objectives
To detect the prevalences of Alloiococcus otitidis, as well as Haemophilus influenzae, Streptococcus pneumoniae, and Moraxella catarrhalis in children with chronic otitis media with effusion (OME) and to simultaneously investigate the colonization of these bacteria in the nasopharynx and palatine tonsils of these patients.
Methods
The study included 34 pediatric patients with OME, and 15 controls without OME. In the study group, A. otitidis, H. influenzae, S. pneumoniae, and M. catarrhalis were investigated in the samples obtained from middle ear effusions (MEE), nasopharyngeal swabs (NPS) and tonsillar swabs (TS), using multiplex polymerase chain reaction (PCR) and conventional culture methods. Only the samples obtained from NPS and TS were studied with the same techniques in the control group.
Go to : 
Chronic otitis media with effusion (OME) is one of the most common diseases of the childhood and it causes acquired hearing loss in children (1). OME is a chronic inflammatory condition and the etiology of inflammation is multifactorial, including viruses, allergy, bacteria and their products, as well as the dysfunction of the Eustachian tube (2). Inflammatory stimuli of infectious agents appear as the most important factor in the development of OME (3). Bacteria can be isolated from the middle ear effusion (MEE) in about 40% of OME patients with conventional culture methods. However, the isolation rate reaches 80% when a molecular method, such as multiplex polymerase chain reaction (PCR) is used (4). Up to date, Haemophilus influenzae, Streptococcus pneumoniae, and Moraxella catarrhalis have been reported as the most common pathogenic microorganisms in OME. Recently, a gram-positive bacterium, Alloiococcus otitidis has been described as an infectious agent that plays a role in OME and acute otitis media. Although identification of this bacterium is difficult in the middle ear effusion by conventional culture methods, A. otitidis was detected in a high proportion of children with OME and acute otitis media by the multiplex PCR method, and its isolation rate was even higher than the three major pathogenic agents H. influenzae, S. pneumoniae, and M. catarrhalis (5-10). A. otitidis and the other three pathogenic agents may colonize in the nasopharynx and tonsils of the OME patients. A. otitidis that colonizes in the tonsils and nasopharynx may cause otitis media with effusion.
In this study, we investigated the presence of A. otitidis, H. influenzae, S. pneumoniae, and M. catarrhalis in the nasopharynx, palatine tonsils and the middle ear effusion, simultaneously, using PCR and cultures in patients with OME. To our knowledge, no studies up to date have investigated, simultaneously, the presence of A. otitidis and other three pathogens in these regions of patients with OME and the colonization of A. otitidis in the palatine tonsils.
Go to : 
The study included 34 pediatric patients with OME (19 females and 15 males aged between 3-16 years with a mean age of 8±3.5 years) and 15 controls subjects. The controls consisted of 9 females and 6 males, without OME, and they were aged between 6-11 years, with a mean age of 8±1.9 years. In addition, they were hospitalized for either septoplasty or frenuloplasty. The children were examined and had surgery in the Department of Otorhinolaryngology in Ankara Training and Research Hospital, in 2007 and 2008. The study was approved by the Ethic Committee of Ankara Training and Research Hospital (No: 0271). Informed consents were obtained from parents of all participating children.
Patients with an effusion lasting more than three months were included in this study. The presence of OME was verified by otomicroscopic examination and tympanometry. They had multiple episodes of infection and had been treated with beta-lactam antibiotics. Eighteen children had bilateral, while 16 children had unilateral disease. The patients did not have any history of recurrent tonsillitis. The control group consisted of 15 children, who did not have any ear disorders or upper respiratory tract infection. None of the children in either group had other known chronic conditions or immunological defects.
MEE samples were obtained by myringotomy, under general anesthesia. The ear canal was cleansed with 10% povidone-iodine before myringotomy. Myringotomy was performed in the anteroinferior part of the tympanic membrane. MEE was aspirated with a specially designed device. The wider side of a sterile 2 mL syringe was connected to the suction tube and an ear suction tube was connected to its thinner side. After the aspirate was collected in the syringe, its thinner side was occluded with an injection pin and its wider side was occluded with a plug. MEE was recorded as mucoid or serous with regard to its physical appearance. Swabs were simultaneously obtained from the nasopharynx (nasopharyngeal swab, NPS) and the palatine tonsils (tonsillar swab, TS). The samples were transported to the microbiology laboratory, within a few hours after their collection to minimize the loss of fastidious microorganisms.
A. otitidis, H. influenzae, S. pneumoniae, and M. catarrhalis were investigated in MEE, NPS, and TS, using multiplex PCR method and conventional cultures in the study group. In patients with bilateral disease, the more effusion in the ear was preferred for MEE sampling. In the control group, only NPS and TS were obtained, while the patients were under general anesthesia, and they were studied with the same technique.
Samples obtained for PCR (half of each MEE sample and one of each swab specimen) were preserved in 1 mL TRIS EDTA buffer at -40℃, until the test was performed. Rest of MEE, along with the second NPS and TS samples were cultured on 5% sheep blood agar, chocolate agar and eosin methylene blue (EMB) agar plates. EMB agar plates were incubated at 37℃ for 24 hours before they were evaluated. Sheep blood agar and chocolate agar plates were incubated in 5% CO2, at 35℃, for 48 hours before their first evaluation. Since A. otitidis is a slow growing bacterium, incubation was prolonged to seven days. Bacterial isolates were identified by standard laboratory procedures. All specimens stored, at -40℃ for molecular tests, were processed by High Pure PCR Template Preparation Kit (Roche, Mannheim, Germany), according to the manufacturer's instructions. The multiplex PCR protocol was performed, as previously described by Hendolin et al. (6). Test was set up in order to detect target DNA regions of A. otitidis, H. influenzae, M. catarrhalis, and S. pneumonia, simultaneously, using specific forward primers for those species, as well as a common lower primer. The multiplex-PCR mixture contained 1.6 µM A. otitidis primer, 1.4 µM H. influenzae primer, 0.2 µM M. catarrhalis primer, 0.04 µM S. pneumoniae primer, 0.4 µM common lower primer, 200 µM dNTP mix, 1X PCR buffer (10 mM Tris-HCl), 1.5 mM MgCl2 and three U Taq Polymerase (Roche) in a reaction volume of 50 µM. The reaction profile was 3 minutes of initial denaturation and 38 cycles at 94℃ for 30 seconds, 66℃ for 45 seconds and 72℃ for one minute, followed by an extension at 72℃ for five minutes. The PCR products were separated in 2% agarose gel, which contained ethidium bromide, and visualized by UV light illumination.
Data were analyzed using SPSS ver. 11.5 (SPSS Inc., Chicago, IL, USA). Data were shown as mean±standard deviation for continuous variables, whereas, categorical data were expressed as the number of the cases and percent. The mean age difference between the groups was compared using Student's t-test. Categorical data were evaluated with Pearson chi-square or Fisher's exact test, where appropriate. A P-value of less than 0.05 was considered statistically significant.
Go to : 
The samples of 34 patients with OME (MEE, NPS, and TS) and the samples of 15 controls (NPS and TS) were analyzed.
A. otitidis was isolated only in MEE of the patients. It was isolated in 12 out of 34 (35%) patients. The bacterium was not isolated in NPS or TS of the patients or the controls. The mean age, gender or effusion type (serous or mucous) were not different with statistical significance between A. otitidis positive and negative cases (P>0.05). Nine out of 12 (75%) A. otitidis positive patients and 20 out of 22 (90.9%) A. otitidis negative patients had adenoid hypertrophy. There was no statistically significant difference between these groups, with regard to the presence of adenoid hypertrophy (P>0.05).
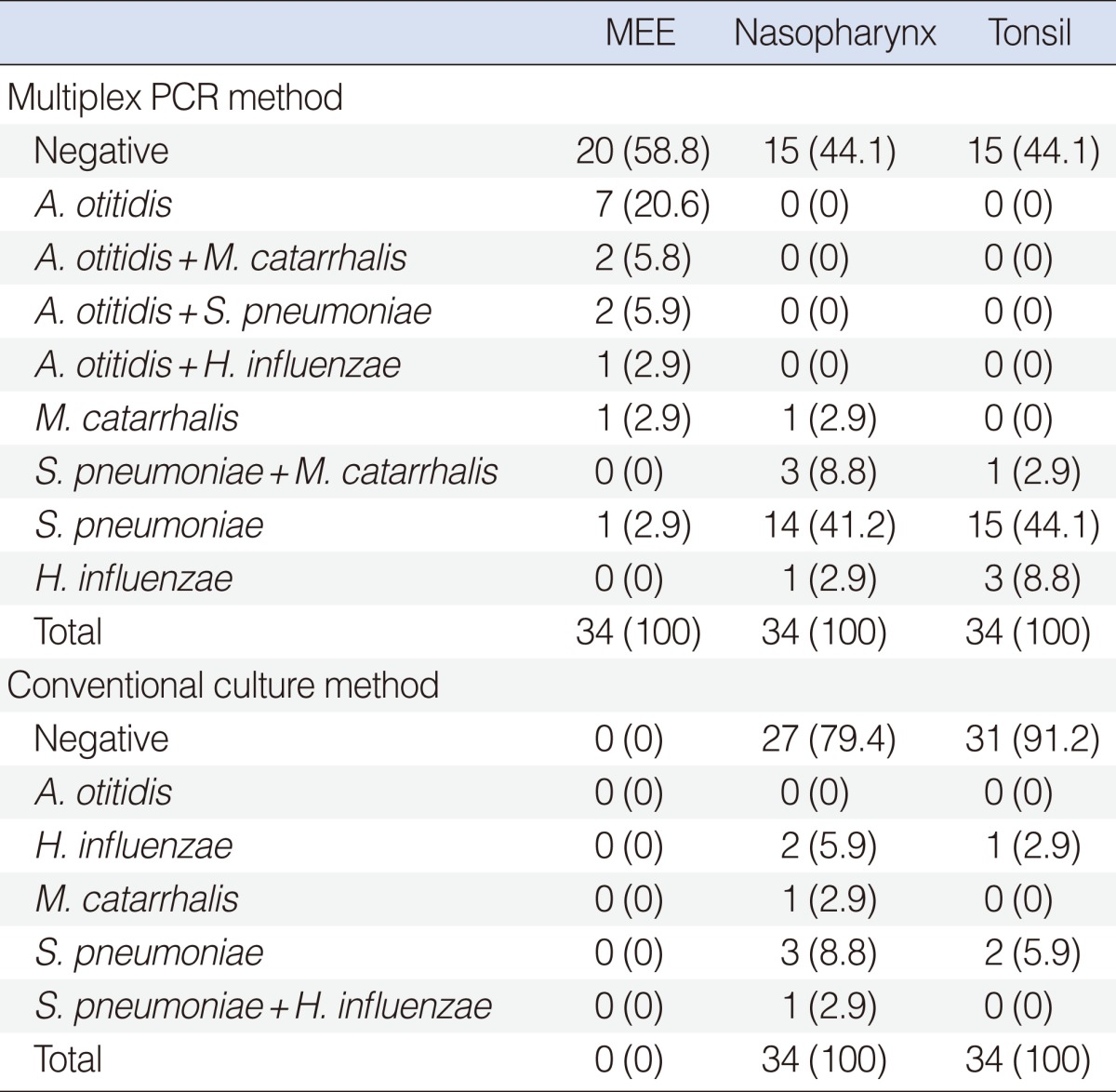
A. otitidis was identified in 12 (35%), S. pneumoniae was identified in 3 (8.8%), M. catarrhalis was identified in 3 (8.8%) and H. influenzae was identified in 1 (2.9%) out of 34 MEE. A. otitidis was identified alone in 7 out of 12 (58%) cases, whereas, it was identified together with one of the other three pathogenic microorganisms in 5 out of 12 (41.7%) cases. No pathogenic agents were isolated in 20 (59%) MEE. S. pneumoniae was detected in 17 (50%), M. catarrhalis was detected in 4 (11.8%) and H. influenzae was detected in 1 (2.9%) out of 34 NPS. S. pneumoniae was detected in 16 (47%), M. catarrhalis was detected in 1 (2.9%), and H. influenzae was detected in 3 (8.8%) out of 34 TS, using the same method. The multiplex PCR method results of the study groups are shown in Table 1.
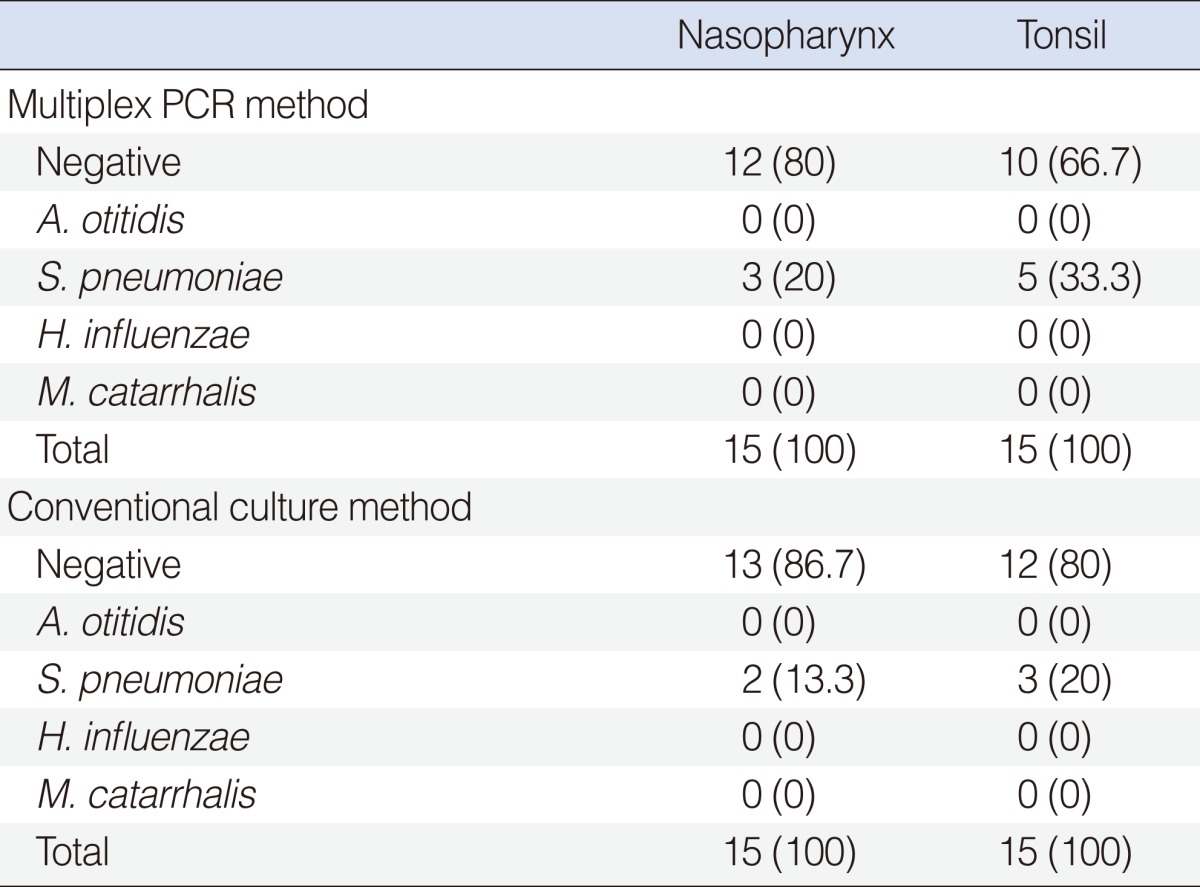
In the control group, S. pneumoniae was detected in 3 (20%) out of the 15 NPS and in 5 (33.3%) out of the 15 TS by multiplex PCR method. A. otitidis or the other two pathogens could not be identified. The results of the control group are shown in Table 2.
A. otitidis was not identified in MEE, NPS or TS samples. Although no bacteria were isolated in MEE cultures, S. pneumoniae was isolated in 4 (11.8%), M. catarrhalis was isolated in 1 (2.9%) and H. influenzae was isolated in 2 (5.9%) out of 34 nasopharyngeal cultures. S. pneumoniae was isolated in 2 (5.9%) and H. influenzae was isolated in 1 (2.9%) out of 34 TS cultures. The culture results of the study group are shown in Table 1.
In the control group, S. pneumoniae was cultured in 2 (13.3%) out of 15 NPS and in 3 (20%) out of 15 TS. Neither A. otitidis nor the other two pathogens could be cultured. The results of the control group are shown in Table 2.
Although A. otitidis was identified in the MEE by multiplex PCR method, it could not be identified by conventional cultures. Additionally, it was not identified either in the NPS or TS, by either the multiplex PCR method or the conventional cultures. Although multiplex PCR method detected A. otitidis and/or one of the other major three pathogens in 14 (41%) out of 34 MEE samples, no pathogenic agents were identified with conventional culture methods.
Go to : 
OME is defined as the presence of fluid in the middle ear, without symptoms or signs of infection, and it is the most common cause of acquired hearing loss in childhood. Thus, it can result in developmental impairment of linguistic, behavioral, motor and social skills (1, 11, 12).
A. otitidis is first recovered from MEE of children with OME by Faden and Dryja (13) in 1989. This bacterium is a weakly catalase-positive, oxidase-negative and strictly aerobic coccus (4). Possible routes of entry of A. otitidis to the middle ear may be through nasopharynx or external ear in patients with perforated tympanic membranes. As well as nasopharynx and external ear, A. otitidis may originate from the upper respiratory tract, such as sinus, nasal cavity, tonsil, pharynx and oral cavity (9). It is difficult to detect A. otitidis in MEE with conventional culture methods, since it grows slowly and requires a special medium to grow (5). A. otitidis has been detected more frequently than the other middle ear pathogens by multiplex PCR (3, 6-8). Previous studies reported that A. otitidis was isolated in 18.5-64% of OME patients (3, 6-10) and in 25-50% of acute otitis media patients (5, 10, 14) with multiplex PCR methods. In the present study, the rate of its isolation was high in children with OME, with a rate of 35%, when studied with multiplex PCR. Further, it was identified more frequently than the other three major pathogenic agents. Our results are in accordance with the literature.
It has been hypothesized that A. otitidis was included in the normal flora of the middle ear cavity. However, previous in vitro and in vivo studies demonstrated that this bacterium had an immunostimulatory capacity, and this indicated that it was not included in the normal flora of the middle ear cavity (15-21). This suggestion was also supported by de Miguel Martinez et al. (12). In their study, A. otitidis was investigated in children with OME and acute otitis media, where the control group had a cochlear implant and did not have otitis for the previous 6 months. The bacterial analysis was performed by means of microbiological culture-specific techniques and the cultures presented negative results in all cases of the control group (12). In our study, A. otitidis was detected alone in 7 out of 12 (58%) cases and it was identified together with one of the other three pathogens in 5 out of 12 (41.7%) cases. We can speculate that A.otitidis is a pathogenic bacterium in the middle ear.
Up to date, apart from the middle ear, A. otitidis was investigated in the outer ear canal (4, 22-24), nasopharynx (4, 9, 22, 24) and maxillary sinus (25). It was most frequently identified in the ear canals of healthy adults (24). Some authors stated that A. otitidis was a commensal inhabitant and is a part of the normal bacterial flora of the human outer ear canal (4, 23, 24).
A. otitidis was investigated in the nasopharynx in a few studies. Durmaz et al. (22) studied 50 healthy children and identified the bacterium in the nasopharynges of 4 children with multiplex PCR. Tano et al. (4) studied 129 patients with upper respiratory tract infection without OME and identified A. otitidis in the nasopharynges out of 9 children, using multiplex PCR. Harimaya et al. (9) showed that nasopharyngeal colonization of A. otitidis increased in otitis-prone children, when compared to those in non-otitis-prone children. Contrary to aforementioned studies, De Baere et al. (24) showed the absence of the bacterium in nasopharynges of 70 healthy adult volunteers. Our study showed that A. otitidis was not present in the nasopharynges of children with OME. However, we detected the bacterium in MEE in 35% of them with multiplex PCR method. Additionally, we did not detect the bacterium in nasopharynges of the control group. These findings suggest that A. otitidis does not colonize in the nasopharynx, and is not included in the normal flora of this region. The discrepancies between the results of this study and the above mentioned studies may depend on different geographic locations and variations in the study protocols, and inclusion criteria among the studies.
To the best of our knowledge, no studies up to date investigated the colonization of A. otitidis in the palatine tonsils of children with OME. We did not identify A. otitidis in the tonsils of study and the control groups.
PCR is a technique that allows for the sensitive and specific detection of bacteria. It is useful for the detection of pathogens that are slowly growing, difficult to culture, or hazardous to handle in a diagnostic laboratory. PCR has been used to improve the sensitivity of bacterial detection (26) and to demonstrate the existence of pathogenic bacterial DNA in culture-sterile effusions in the middle ear from pediatric patients (27). There were some clinical and experimental studies, which suggest that viable, intact bacteria were required for the detection of bacterial DNA by PCR-based assays (10, 28).
Some authors showed that multiplex PCR methods were more useful than that of the cultures for identifying A. Otitidis (6-8). In accordance with these studies, we observed in our study that multiplex PCR was more useful than that of the conventional cultures for detecting A. otitidis and other three major pathogens.
In Leskinen et al. (7)'s study, A. otitidis was detected more frequently with greater significance in mucous effusions, which represent a more chronic stage of otitis media, when compared to mucoserous effusions. However, we did not find any relationship between the presence of A. otitidis and the effusion type, in our study.
de Miguel Martinez and Macias (12) reported that A. otitidis was sensitive to ampicillin and cefotaxime, but it was resistant to macrolides and cotrimoxazol. Harimaya et al. (5) reported that A. otitidis was frequently detected in patients who received a beta-lactam antibiotics or erythromycin. In our study, although all cases had received beta-lactam antibiotics for more than three months, A. otitidis was identified in 35% of the patients with OME. In light of these findings, we supposed that A. otitidis might be resistant to beta-lactams.
In conclusion, we showed that A. otitidis was frequently isolated in MEE of children with OME, when studied with multiplex PCR. However, it could not be identified in the nasopharynx or tonsils of the study and either with the control groups with multiplex PCR or conventional cultures. This study results showed that the prevalence of A. otitidis was high in children with OME. Our results also revealed that A. otitidis did not colonize in the nasopharynx or in the tonsil, and it was not included in the normal flora of these regions. New, larger studies are needed to further clarify these issues.
Go to : 
Notes
This report was presented at the meeting (32. Türk Ulusal KBB ve BBC kongresi 27-31 October 2010, Antalya, Turkey).
Go to : 
References
1. Serbetcioglu B, Ugurtay O, Kirkim G, Mutlu B. No association between hearing loss due to bilateral otitis media with effusion and Denver-II test results in preschool children. Int J Pediatr Otorhinolaryngol. 2008; 2. 72(2):215–222. PMID: 18045700.

2. Keles B, Ozturk K, Arbag H, Gunel E, Ozer B. Frequency of pharyngeal reflux in children with adenoid hyperplasia. Int J Pediatr Otorhinolaryngol. 2005; 8. 69(8):1103–1107. PMID: 16005352.

3. Guvenc MG, Midilli K, Inci E, Kuskucu M, Tahamiler R, Ozergil E, et al. Lack of Chlamydophila pneumoniae and predominance of Alloiococcus otitidis in middle ear fluids of children with otitis media with effusion. Auris Nasus Larynx. 2010; 6. 37(3):269–273. PMID: 19879704.

4. Tano K, von Essen R, Eriksson PO, Sjostedt A. Alloiococcus otitidis-otitis media pathogen or normal bacterial flora? APMIS. 2008; 9. 116(9):785–790. PMID: 19024598.

5. Harimaya A, Takada R, Hendolin PH, Fujii N, Ylikoski J, Himi T. High incidence of Alloiococcus otitidis in children with otitis media, despite treatment with antibiotics. J Clin Microbiol. 2006; 3. 44(3):946–949. PMID: 16517881.
6. Hendolin PH, Markkanen A, Ylikoski J, Wahlfors JJ. Use of multiplex PCR for simultaneous detection of four bacterial species in middle ear effusions. J Clin Microbiol. 1997; 11. 35(11):2854–2858. PMID: 9350746.

7. Leskinen K, Hendolin P, Virolainen-Julkunen A, Ylikoski J, Jero J. The clinical role of Alloiococcus otitidis in otitis media with effusion. Int J Pediatr Otorhinolaryngol. 2002; 10. 66(1):41–48. PMID: 12363421.

8. Kalcioglu MT, Oncel S, Durmaz R, Otlu B, Miman MC, Ozturan O. Bacterial etiology of otitis media with effusion: focusing on the high positivity of Alloiococcus otitidis. New Microbiol. 2002; 1. 25(1):31–35. PMID: 11837388.
9. Harimaya A, Takada R, Somekawa Y, Fujii N, Himi T. High frequency of Alloiococcus otitidis in the nasopharynx and in the middle ear cavity of otitis-prone children. Int J Pediatr Otorhinolaryngol. 2006; 6. 70(6):1009–1014. PMID: 16310863.

10. Kaur R, Adlowitz DG, Casey JR, Zeng M, Pichichero ME. Simultaneous assay for four bacterial species including Alloiococcus otitidis using multiplex-PCR in children with culture negative acute otitis media. Pediatr Infect Dis J. 2010; 8. 29(8):741–745. PMID: 20335823.

11. Eser OK, Ipci K, Alp S, Akyol U, Unal OF, Hascelik G, et al. Efficacy of nasopharyngeal culture in identification of pathogen in middle ear fluid in chronic otitis media with effusion. Indian J Med Microbiol. 2009; Jul-Sep. 27(3):237–241. PMID: 19584505.

12. de Miguel Martinez I, Macias AR. Serous otitis media in children: implication of Alloiococcus otitidis. Otol Neurotol. 2008; 6. 29(4):526–530. PMID: 18418283.
13. Faden H, Dryja D. Recovery of a unique bacterial organism in human middle ear fluid and its possible role in chronic otitis media. J Clin Microbiol. 1989; 11. 27(11):2488–2491. PMID: 2808673.

14. Leskinen K, Hendolin P, Virolainen-Julkunen A, Ylikoski J, Jero J. Alloiococcus otitidis in acute otitis media. Int J Pediatr Otorhinolaryngol. 2004; 1. 68(1):51–56. PMID: 14687687.

15. Tarkkanen J, Himi T, Harimaya A, Carlson P, Ylikoski J, Mattila PS. Stimulation of adenoidal lymphocytes by Alloiococcus otitidis. Ann Otol Rhinol Laryngol. 2000; 10. 109(10 Pt 1):958–964. PMID: 11051437.

16. Kita H, Himi T, Fujii N, Ylikoski J. Interleukin-8 secretion of human epithelial and monocytic cell lines induced by middle ear pathogens. Microbiol Immunol. 2000; 44(6):511–517. PMID: 10941934.

17. Himi T, Kita H, Mitsuzawa H, Harimaya A, Tarkkanen J, Hendolin P, et al. Effect of Alloiococcus otitidis and three pathogens of otitis media in production of interleukin-12 by human monocyte cell line. FEMS Immunol Med Microbiol. 2000; 10. 29(2):101–106. PMID: 11024348.
18. Harimaya A, Himi T, Fujii N, Tarkkanen J, Carlson P, Ylikoski J, et al. Induction of CD69 expression and Th1 cytokines release from human peripheral blood lymphocytes after in vitro stimulation with Alloiococcus otitidis and three middle ear pathogens. FEMS Immunol Med Microbiol. 2005; 3. 43(3):385–392. PMID: 15708312.
19. Harimaya A, Takada R, Himi T, Yokota S, Fujii N. Evidence of local antibody response against Alloiococcus otitidis in the middle ear cavity of children with otitis media. FEMS Immunol Med Microbiol. 2007; 2. 49(1):41–45. PMID: 17094788.
20. Harimaya A, Fujii N, Himi T. Preliminary study of proinflammatory cytokines and chemokines in the middle ear of acute otitis media due to Alloiococcus otitidis. Int J Pediatr Otorhinolaryngol. 2009; 5. 73(5):677–680. PMID: 19185927.

21. Konishi M, Nishitani C, Mitsuzawa H, Shimizu T, Sano H, Harimaya A, et al. Alloiococcus otitidis is a ligand for collectins and Toll-like receptor 2, and its phagocytosis is enhanced by collectins. Eur J Immunol. 2006; 6. 36(6):1527–1536. PMID: 16708401.

22. Durmaz R, Ozerol IH, Kalcioglu MT, Oncel S, Otlu B, Direkel S, et al. Detection of Alloiococcus otitidis in the nasopharynx and in the outer ear canal. New Microbiol. 2002; 4. 25(2):265–268. PMID: 12019737.
23. Frank DN, Spiegelman GB, Davis W, Wagner E, Lyons E, Pace NR. Culture-independent molecular analysis of microbial constituents of the healthy human outer ear. J Clin Microbiol. 2003; 1. 41(1):295–303. PMID: 12517864.

24. De Baere T, Vaneechoutte M, Deschaght P, Huyghe J, Dhooge I. The prevalence of middle ear pathogens in the outer ear canal and the nasopharyngeal cavity of healthy young adults. Clin Microbiol Infect. 2010; 7. 16(7):1031–1035. PMID: 19895585.

25. Kalcioglu MT, Durmaz B, Aktas E, Ozturan O, Durmaz R. Bacteriology of chronic maxillary sinusitis and normal maxillary sinuses: using culture and multiplex polymerase chain reaction. Am J Rhinol. 2003; May-Jun. 17(3):143–147. PMID: 12862402.

26. Hendolin PH, Paulin L, Ylikoski J. Clinically applicable multiplex PCR for four middle ear pathogens. J Clin Microbiol. 2000; 1. 38(1):125–132. PMID: 10618075.

27. Post JC, Preston RA, Aul JJ, Larkins-Pettigrew M, Rydquist-White J, Anderson KW, et al. Molecular analysis of bacterial pathogens in otitis media with effusion. JAMA. 1995; 5. 273(20):1598–1604. PMID: 7745773.

28. Post JC, Aul JJ, White GJ, Wadowsky RM, Zavoral T, Tabari R, et al. PCR-based detection of bacterial DNA after antimicrobial treatment is indicative of persistent, viable bacteria in the chinchilla model of otitis media. Am J Otolaryngol. 1996; Mar-Apr. 17(2):106–111. PMID: 8820185.

Go to : 




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download