INTRODUCTION
MATERIALS AND METHODS
Study subjects
Table 1.
| Characteristics | AR (n=39) | Non-AR (n=22) | P-value |
|---|---|---|---|
| Sex | 0.415 | ||
| Male | 26 | 12 | |
| Female | 13 | 10 | |
| Diagnosis | 0.587 | ||
| CRSsNP | 25 | 12 | |
| CRSwNP | 14 | 10 | |
| Age (yr) | 43.54 ±17.45 | 36.14 ±20.68 | 0.142 |
| Serum eosinophils count (cells/µL) | 314.8 ±293.9 | 153.7 ±94.81 | 0.006* |
| Serum ECP (µg/L) | 25.57 ±31.70 | 15.69 ±13.39 | 0.141 |
| Serum total IgE (U/mL) | 277.4 ±440.41 | 42.22 ±67.76 | <0.001** |
HRV-16 infection in organ culture
Determination of successful HRV infection
PD-L1, PD-L2, and cytokine mRNA in the nasal mucosa
PD-L1, PD-L2, ICAM-1, and IL-10 protein expression in the nasal mucosa
Statistical analysis
RESULTS
HRV infection in the allergy and non-allergy groups
Different responses to HRV infection according to the presence or absence of AR
Expression of PD-L1 and PD-L2 in the nasal mucosa
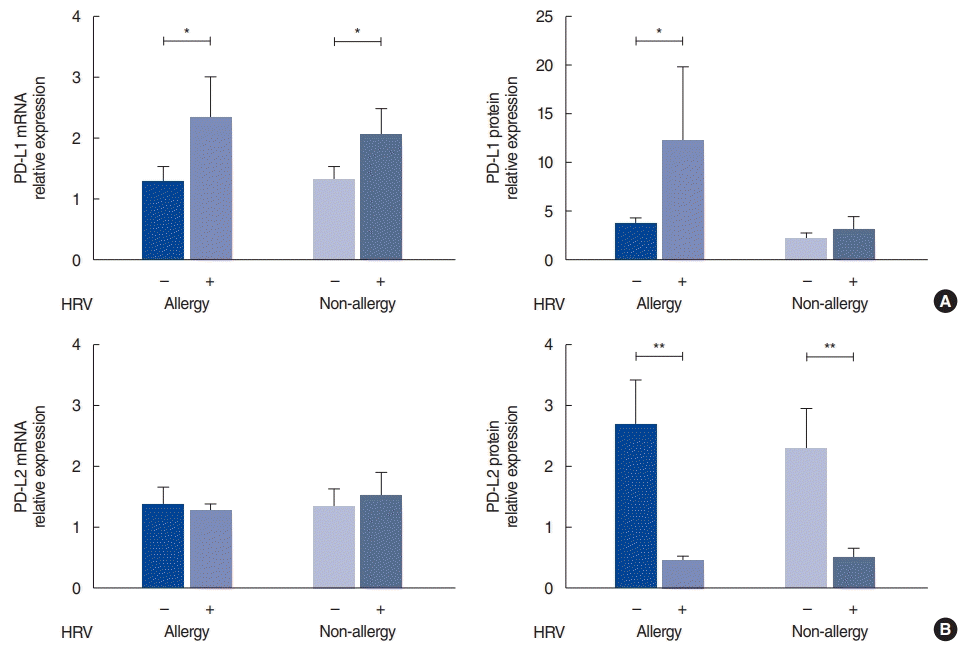 | Fig. 1.Programmed cell death-ligand (PD-L)1 and PD-L2 expression after human rhinovirus (HRV) infection in the allergy and non-allergy groups. (A) mRNA and protein levels of PD-L1 after HRV infection (B) mRNA and protein levels of PD-L2 after HRV infection. The relative expression of mRNA was calculated based on the ∆∆CT method; the relative expression of protein represents the ratio of PD-L1 or PD-L2 to β-actin. Values are presented as mean±standard error of the mean.
*P<0.05, **P<0.001.
|
Expression of ICAM-1 in the nasal mucosa
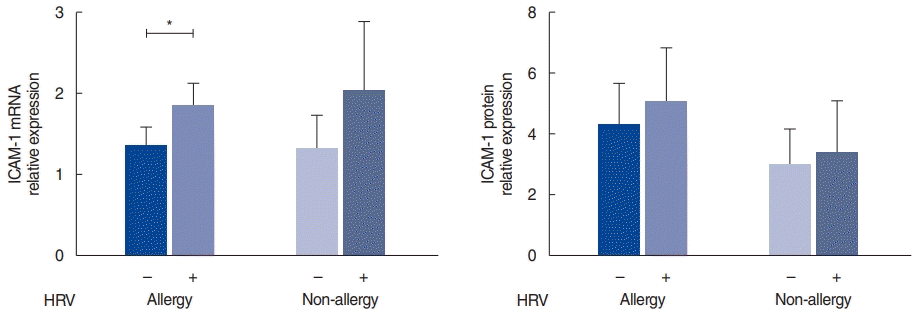 | Fig. 2.Intracellular adhesion molecule 1 (ICAM-1) expression after human rhinovirus (HRV) infection in the allergy and non-allergy groups. mRNA and protein levels of ICAM-1 after HRV infection. The relative expression of mRNA was calculated based on the ∆∆CT method; the relative expression of protein represents the ratio of ICAM-1 to β-actin. Values are presented as mean±standard error of the mean.
*P<0.05.
|
Expression of IL-4, IL-5, IL-10, and IFN-γ in the nasal mucosa
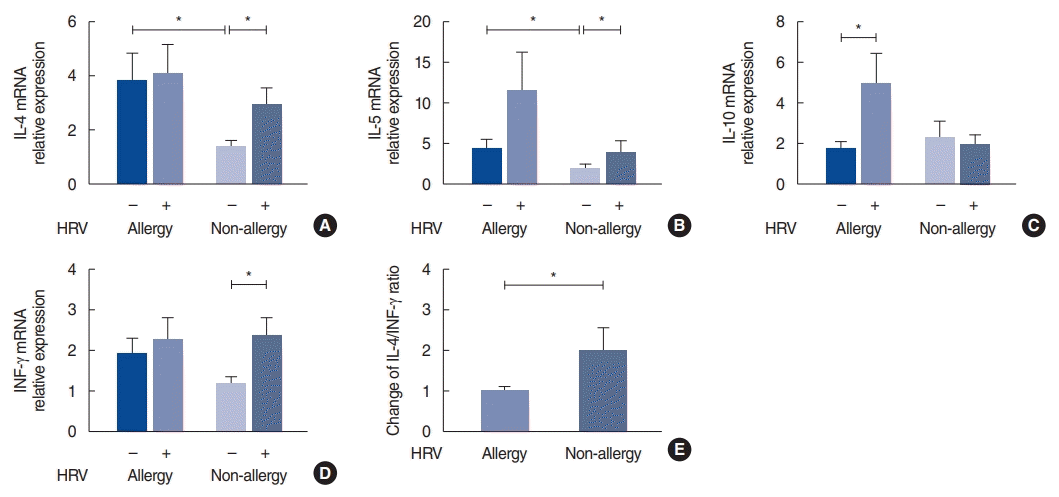 | Fig. 3.Interleukin (IL)-4, IL-5, IL-10 and interferon (IFN) levels after human rhinovirus (HRV) infection in the allergy and non-allergy groups. (A) IL-4 mRNA, (B) IL-5 mRNA, (C) IL-10 mRNA, (D) IFN-γ mRNA levels after HRV infection. (E) Change in the IL-4/INF-γ ratio after HRV infection in each subject. The relative expression of mRNA was calculated based on the ∆∆CT method. Values are presented as the mean±standard error of the mean.
*P<0.05.
|
Association between PD-L1 and IL-10 in the nasal mucosa
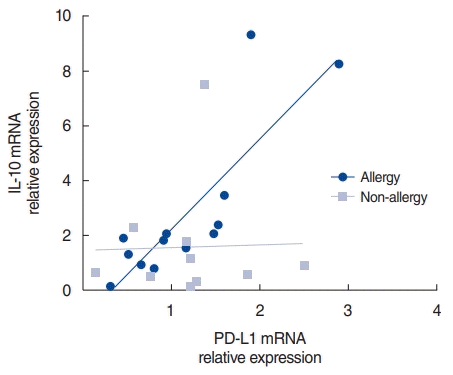 | Fig. 4.Correlation of programmed cell death-ligand (PD-L)1 and interleukin (IL)-10 after human rhinovirus (HRV) infection. PD-L1 and IL-10 mRNA levels after HRV infection in the allergy and non-allergy groups. The relative expression was calculated as the ratio of the expression in the infected to control tissue in each individual. |




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download