Abstract
Biosignals such as electrocardiogram or photoplethysmogram are widely used for determining and monitoring the medical condition of patients. It was recently discovered that more information could be gathered from biosignals by applying artificial intelligence (AI). At present, one of the most impactful advancements in AI is deep learning. Deep learning-based models can extract important features from raw data without feature engineering by humans, provided the amount of data is sufficient. This AI-enabled feature presents opportunities to obtain latent information that may be used as a digital biomarker for detecting or predicting a clinical outcome or event without further invasive evaluation. However, the black box model of deep learning is difficult to understand for clinicians familiar with a conventional method of analysis of biosignals. A basic knowledge of AI and machine learning is required for the clinicians to properly interpret the extracted information and to adopt it in clinical practice. This review covers the basics of AI and machine learning, and the feasibility of their application to real-life situations by clinicians in the near future.
Biosignals are signals such as electrocardiogram (ECG), electroencephalogram (EEG), and photoplethysmogram (PPG) that are obtained from patients. Monitoring devices usually keep a check on the real-time status of patients in the intensive care unit (ICU) as well as those who have undergone surgery, for the entire duration of treatment; measurements like ECG or electromyogram are widely used even in out-patient based practices as they are non-invasive, and provide valuable information.
Information in biosignals is usually not expressed directly and diverse data are hidden in signal waveforms. For example, it is well known that variation in the time interval between heartbeats or heart-rate variability that can be extracted from an ECG or PPG is associated with mortality or diverse unfavorable clinical outcomes [1,2]. Another example is the QT interval in an ECG that represents the time interval between the start of ventricular contraction to end of repolarization of the heart muscle. Some drugs prolong the QT interval by prohibiting the repolarization process of the heart muscle; however, extended prolongation could cause life-threatening arrhythmia known as torsade de points [3,4]. Therefore, drugs having the potential for QT prolongation were withdrawn from the market [5]. P-wave indices that reflect the status of the atrium are another example; recent study reported that additional information of P-wave indices along with the CHA₂DS₂-VASc score can predict the probability of an ischemic stroke in atrial fibrillation patients more accurately than on the basis of the CHA₂DS₂-VASc score alone [6].
Recent advances in artificial intelligence (AI) provide opportunities to reveal hidden information in biosignals that is not apparent using conventional methods of analysis. One of the current studies reported that an AI-enabled algorithm was able to identify patients with atrial fibrillation although their ECG signals were in sinus rhythm [7], implying that there are some indications facilitating the AI model to identify an alteration of the atrium from the ECG. Another recent study discovered that the ejection fraction of the heart could be determined from an ECG and a cardiac contractile dysfunction could be detected by applying the AI model to an ECG signal [8]. From EEG signals, it was possible to predict sex when deep learning was applied to brain rhythms [9]. Another study reported that information on what subjects were viewing could be obtained from an EEG and an AI model could reconstruct images from the EEG measured while the subjects were viewing the images [10]. These studies showed that only a small part of information from biosignals is actually utilized in clinical practice (Fig. 1). However, it must be noted that the morphology of features extracted by or used in AI models are remarkably different from those that are widely used. AI-enabled features are usually in a black box i.e., they are invisible, indescribable, and indefinable by human perception whereas conventional features are clearly visible and easily defined by simple rules. Therefore, to use AI-enabled features suitably in real-world practice, clinicians must know and understand the working of an AI model to extract hidden information from largely stacked data.
To enable clinicians to understand and use AI-enabled features from biosignals, this study presents the following: 1) a brief introduction of AI and machine learning for clinicians, 2) recent studies that demonstrate the feasibility of utilizing AI-enabled features from biosignals for clinical use, and 3) available databases to conduct AI-based biosignal research.
AI refers to any computer software that can mimic cognitive functions or human intelligence. However, as the real mechanism of human cognitive functions has not yet been discovered, it is impossible to develop software that follows the mechanism of real intelligence; instead, scientists strategically developed software that appears to have cognitive functions regardless of its underlying mechanism.
AI is a concept with diverse subcategories defined by various principles (Fig. 2), classified according to whether it learns patterns from data or not. An example of AI that does not depend on data is the rule-based system designed by domain experts to handle specific problems well fitted to a hypothesis. In this case, the level of knowledge of the experts is important rather than the amount of data; however, this system is rigid and fails if there is a variation in the problem. The other approach called machine learning is to determine certain patterns from the data. This allows the software to change certain parts such as weights or coefficients in accordance with the inputted data.
Machine learning can be classified into three categories: The first is supervised learning for which two datasets must be prepared namely, input data for pattern training and information labels regarding the category or value of the input data (for example, normal vs cancer or survival vs death). Based on the information labels, the supervised learning algorithm tries to determine a pattern where the output is a category (classification problem) or real value (regression problem). By contrast, unsupervised learning is an algorithm that identifies inherent patterns in the data itself without labeled information. For example, clustering algorithms such as k-means or hierarchical clustering divides each data point into groups (usually the number of groups is predetermined by researcher) based on the distribution. Finally, reinforcement learning aims to set the best policy to solve a certain problem within the computationally designed space by trial and error.
Deep learning, the most advanced form of artificial neural networks, is an architecture that allows diverse methods to fit models to a given data by learning patterns in the data. It can be used for supervised learning, unsupervised learning, as well as reinforcement learning.
Deep learning is a part of machine learning; however, it has distinct characteristics as compared to traditional machine learning algorithms. First, in traditional machine learning, it is crucial to extract important features from the raw data by using domain knowledge; however, deep learning takes raw data as the input, extracts the features within the data, and learns the patterns by itself. Deep learning requires basic preprocessing such as normalization or noise removal; however, it lowers the barrier of entry for specialized domains of data, as it is relatively free from feature engineering based on domain knowledge. For example, suppose we need to develop a model that can distinguish between chest x-ray images of normal lungs and those afflicted by cancer. In earlier machine learning, the researcher had to extract features such as the shape of the nodule or distribution of colors that could be observed in images, and input that to the model. However, deep learning can directly extract the information that is relevant for discriminating between the two. Second, in traditional machine learning, the performance is usually restricted due to the limited predefined set of features and does not improve regardless of the amount of data used for training whereas deep learning requires a larger amount of data as the direct feature extraction process by researchers is omitted. The deep learning model can identify patterns clearly, when more data is added; hence, the diversity, clearance, and usefulness of the extracted features are enhanced leading to an improvement in the performance. Third, traditional machine learning provides the significance of the used features and information that have a major role whereas deep learning is difficult to interpret as it is similar to a black box. This may prove to be a barrier in adopting it for medical applications; however, it is now possible to understand deep learning indirectly through algorithms such as Gradient-weighted Class Activation Mapping [11]. This has led to an increased possibility of its use in the medical field.
Deep learning was particularly useful in visual problem solving in the early stages of its development. In image classification problems like ImageNet datasets, the convolutional neural network (CNN), one of several types of deep learning, showed an accuracy greater than 95% [12]. The CNN extracts features from images through filters that derive the morphological features from a small part (called window size) in an image. The CNN filter uses an element-wise product and weights, and the features from the whole image are analyzed by moving it (Fig. 3A). Learning through a CNN involves finding the appropriate filters and their weights to classify labels. For example, given that the shape of a nodule is a key feature to classify it as malignant or benign, a filter randomly initialized in the first stage cannot react to the specific shape; however, the CNN weights of the filter are updated after training to yield a strong response. The result extracted through the filter is called the feature map. It is possible to derive more abstracted features from the images when the CNN is applied deeply.
The deep learning designed to analyze time series data is the recursive neural network (RNN). Time series data usually have an autocorrelation where the previous value affects the next. To grasp the pattern of such time series data, it is necessary to reflect this autocorrelation in deep learning, for which both the past and current data must be considered. When the RNN analyzes the result in the Nth time step, the result is derived from the (N−1)-th time step as an input value. At this time, the output of the Nth time step is delivered to the next step i.e., (N+1)-th step. The information transferred across the time steps is called a hidden state, which is expected to remember the information of the previous time steps. From the above explanation, it appears that different neural networks are needed for every time step; however, the output from the RNN is transferred back to the same neural network (Fig. 3B). This is why it is referred to as an RNN.
A drawback in the basic RNN model is that the information of past data becomes blurred as the time step length of the analysis data increases. To address this, there are improved models such as long short-term memory [13] and gated recurrent neural network [14], which add a separate neural network to carry the information from past data. In current studies, RNN-based models are also applied with an attention mechanism [15] to provide clues that identify relevant context [12,16,17].
The autoencoder is another type of artificial neural network architecture consisting of an encoder that compresses the original raw data to a smaller size and decoder that restores the compressed information into original data (Fig. 4). It is classified as unsupervised learning as only original data is needed, without label information. It is also called representation learning in terms of identifying the intrinsic structure of data and extracting features from them. Identifying key information and compressing it into smaller dimensions is similar to the traditional methods of reduction of dimensions such as principal component analysis; however, the autoencoder has the advantage of representing more complex data spaces through non-linear functions.
The autoencoder tries to learn to minimize the reconstruction error that represents the difference between the original data X and reconstructed data X ', which is the output from the decoder. The information compressed by the encoder is the latent variable (h). If the original data is restored with the latent variable of a size smaller than the original data, it indicates that the encoder identified the pattern or internal structure of the data and extracted the main features. In such a case, the latent variable extracted by the encoder can be used as input data for other machine learning models like clustering or classification (Figs. 4B and 5).
h= fencoder (X)
X'=fdecoder (h)
Reconstruction Error(Mean Squared Error)=‖X-X'‖2
As aforedescribed, deep learning is capable of finding patterns in complex data that are difficult for humans to recognize, and abstracting them into smaller data. To solve problems with less complexity, the existing statistical models or traditional machine learning also provides satisfactory results. However, for analysis of complex data, where the hidden features are abstract and difficult to extract, deep learning which can extract and utilize more diverse hidden information shows more accurate .
Recent advances in computational power and the concurrent rapid increment of data generated in healthcare systems have encouraged researchers to apply AI for assisting various practices in hospitals and other healthcare institutions. As reviewed in the previous section, AI can detect meaningful information in a dataset and recent studies have shown the superiority of AI over human ability in detecting diseases, predicting results, and deciding treatment policies. Some of the AI applications are outlined below:
Introduction of AI in medicine enables the analysis of high-density data such as biosignals; in particular, the conventional 12-electrode ECG is at the forefront of this research. One example is a study that screens for hyperkalemia (serum potassium levels > 5.5 mEq/L) among patients with chronic kidney disease based on a 12-electrode ECG [18]. In the study, researchers used data from 1,576,581 ECGs collected from a single hospital in the United States from 449,380 patients with potassium test records taken within 12 hours of recording their ECG. The deep CNN using the data from ECG leads I and II among 12 leads showed an area under the curve (AUC) of 0.88 for hyperkalemia. An external validation conducted with datasets from other hospitals in the United States also showed good results with an AUC greater than 0.85. Hyperkalemia was previously known to change certain patterns in ECGs such as a tented P wave and sine wave; however, there was a controversy regarding its significance in real practice [19]. The study showed that AI can determine hyperkalemia in a non-invasive way without a blood test and can contribute to the monitoring of abnormalities of electrolytes in patients with chronic kidney disease. In addition, studies on the application of AI to detect and classify arrhythmias such as detecting atrial fibrillation [20] or classifying myocardial infarction [21] using 12-electrode ECG databases are regularly carried out and the results demonstrate an efficacy as high as or higher than that of a human cardiologist.
Recent studies are not limited to the detection of arrhythmias at the time of measurement of ECGs. The introduction of deep learning such as CNN in ECG research has enabled computers to detect even the smallest changes that traditional ECG analysis was unable to interpret. As a result, it is possible to predict arrhythmias not only during the measurement of an ECG but also the possibility of its occurrence in future or anticipate the results of more invasive or expensive tests. For example, a deep learning model recently trained by using the 12-electrode ECG could predict whether atrial fibrillation would occur within a month for patients with a normal sinus rhythm in their ECG obtained by the conventional method [7]. From the 180,922 patients in the period 1993–2017 in the United States, 649,931 ECGs which showed a normal sinus rhythm were used for analysis. The model was trained to detect ECG patterns that could be observed before the first occurrence of atrial fibrillation and predicted whether atrial fibrillation would occur or not. The prediction was reliable with an AUC of 0.87, sensitivity of 0.79, specificity of 0.80, F1 score of 0.39, and overall accuracy of 0.79. This study suggests that deep learning can detect minor changes that are not visually confirmed before the onset of arrhythmia.
Another study by the same research group developed a deep learning model that could predict cardiac contractile dysfunction (ejection fraction less than 35%) using the ECGs of 44,959 patients measured in a period of two weeks before and after their echocardiography [8]. The results of the prediction showed an AUC of 0.93, sensitivity of 0.86, specificity of 0.86, and accuracy of 0.86. The risk ratio for cardiac contractile dysfunction of patients, who were classified as low ejection fraction patients by AI but did not have cardiac contractile dysfunction in the ultrasound at the time of prediction, was estimated to be four times higher as compared to those patients who were classified as normal when the Cox regression analysis was performed.
Another example is the use of ECG rhythms recorded by automated external defibrillators during chest compressions in 1151 cases of cardiac arrest [22]. In the study, the AI model could predict a functionally intact survival rate after acute cardiac arrest. The performance was not highly satisfactory as the AUC was 0.75; however, it showed the possibility of a novel use of ECG and suggested the establishment of a precise cardiopulmonary resuscitation guide based on this.
Other types of biosignals were also used in studies related to AI. For example, in the analysis of frontal EEG measurements of 174 patients in ICU, a deep learning model could predict the level of consciousness (AUC of 0.70) and delirium with (AUC of 0.80). An EEG signal can be used for epileptic focus localization [23], emotion classification [24], and detection of psychotic disorders such as unipolar depression [25] with high reliability, if AI is adopted.
These developments can also be applied to patients outside a hospital, particularly the newly generated data observed on wearable devices that are supported by industry and are of considerable medical value. In a recent study, the deep learning model used 91,232 single lead ECGs of 53,549 patients to successfully categorize the raw input ECG signal into 12 different rhythms without any preprocessing [26]. The averaged AUC of 0.98 was superior to the average reading by board-certified practicing physicians.
Studies using non-medical devices such as smart watches or other commercial wearable devices are also actively underway. A study was conducted recently in the United States on 419,297 members with no history of atrial fibrillation who agreed to being monitored by a smartphone (Apple iPhone) app [27]. The study showed that 84% of the notifications from the irregular pulse notification algorithm were concordant with atrial fibrillation.
In addition to ECG, wearable devices are also useful to monitor the level of activity of people. A study that analyzed data from 2354 prospective cohorts in the United States confirmed the relationship between activity and brain volume, indicating that mild exercise is associated with larger brain volume [28]. An additional one hour of mild exercise in a day can reduce the age of the brain by 1.1 years. Another study tracked the activity of 95 patients with multiple sclerosis with a commercial wearable device for one year [29] wherein the less active group showed a worsening of clinical prognosis.
The increase in the number of commercially available wearable devices is expected to contribute to the collection of large amounts of biosignals from the general population rather than patients alone. It will become a valuable source of data for developing an AI model for cost-effective screening in the pre-hospital stage. Further, it may increase patient compliance by allowing them to participate in their treatment.
CDSSs are computer systems designed to support the decision of a clinician regarding individual patients. The primary purpose is to reduce errors by clinicians, such as prescribing drugs that may react adversely with those being ingested by patients or failing to conduct crucial tests. These traditional functions follow rule-based algorithms designed by medical experts. However, if AI is adopted in a CDSS, it can provide additional valuable information to increase the efficiency and quality of regular practice by predicting diverse clinical events such as mortality with reliable accuracy. Moreover, it can prove to be a novel digital biomarker that is valuable to make next-step decisions that the clinician might have missed.
The EWS is a similar system to aid the decision of the physician, though the focus is on the prompt notification of impending risk to ensure timely management to reduce the consequences of an adverse event. The algorithm used in current EWSs is based on simple rules; therefore, the accuracy is low and applicable area is limited. However, the ability of AI to extract valuable information not easily known to humans provides timely notification with greater accuracy and wider range of applicable areas.
These systems for supporting clinicians provide an opportunity to ensure high quality practice at low cost in advanced hospital care. However, they are based on existing medical records that are limited in detailed information compared to biosignal data. Applying AI to these systems based on biosignals can solve these problems as the number of hospitals that have systems for automatically recording continuous biosignals is increasing. The real-time biosignal recording systems will enable to fill the gap in information in regular medical records required for CDSSs or EWSs.
SaMD is software that can be used for medical purposes independent of a hardware medical device. Traditionally, software is a part of hardware; however, owing to advanced AI technology it is now a type of medical device supporting the clinician. AI models, such as diagnostic software for detecting diseases through radiologic images or ECGs, that can detect malignant nodules or cardiovascular disease are considered to be SaMDs. The US Food and Drug Administration (FDA) and Korea FDA have prepared the management and regulation of this software as a medical device [30,31].
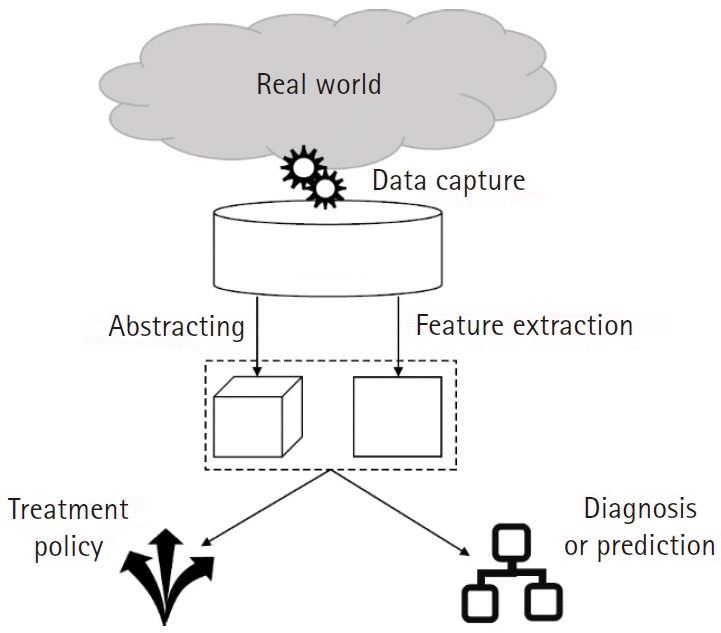
AI application processes in the medical field based on biosignal data begin with the collection of large amounts of raw data (Fig. 5). Several previous studies have developed biosignal data collection systems in hospitals and some of these have made significant contributions to the field of biosignal data research as given below:
MIMIC-III is a database collected from more than 40,000 patients in the ICU at Beth Israel Deaconess Medical Center in the United States from 2001 to 2012 [32]. It consists of demographics, biosignal measurements (including waveform data for some subsets), laboratory test results, procedures, drugs, caregiver notes, imaging reports, and mortality (in and out of the hospital). This large database is freely available worldwide and is the most popular database in biosignal research.
eICU database is one that includes data from 200,859 patients in 335 units of 208 hospitals in the United States from 2014 to 2015 [33]. It consists of biosignal measurements (vital signs but not waveforms), laboratory test results, drugs, acute physiology and chronic health evaluation (APACHE) components, care plan information, diagnostics, patient history, and treatment. It is publicly available and managed by the same research group as the MIMIC-III database.
The Ajou University Hospital Biosignal database contains data of patients who were in the ICUs or emergency rooms of the Ajou University Hospital in Korea since August 2016 [34]. Until November 2019, this large database consisted of more than 19,000 patients with all the biosignal waveform data and vital signs during the period of the patients' admission to discharge from the hospital. All the biosignal data were linked with the electronic medical record data. An automated system collected data from approximately 140 bedside patient monitors continuously. The database is not publicly available yet; however, data access is possible by conducting collaborative work with the research team.
VitalDB database is a biosignal database consisting of data from 6388 patients who underwent surgery in 10 operating rooms at Seoul National University Hospital from 2016 to 2019 [35]. It includes biosignal waveform data, vital signs during surgery, various clinical parameters related to surgery, and laboratory test results.
The rapid advancement in AI technology presents an opportunity to increase the usefulness of biosignals. It can be used not only to increase the accuracy in detecting critically important events but also to predict events or suggest further treatment by extracting valuable hidden information in biosignals. AI based on biosignals is a powerful platform to support daily clinical practice in hospitals and healthcare in daily life.
Notes
Author Contributions
Dukyong Yoon (Conceptualization; Supervision; Visualization; Writing – original draft; Writing – review & editing)
Jong-Hwan Jang (Writing – original draft)
Byung Jin Choi (Writing – original draft)
Tae Young Kim (Writing – original draft)
Chang Ho Han (Writing – original draft)
References
1. Zhou X, Ma Z, Zhang L, Zhou S, Wang J, Wang B, et al. Heart rate variability in the prediction of survival in patients with cancer: a systematic review and meta-analysis. J Psychosom Res. 2016; 89:20–5.

2. Rajendra Acharya U, Paul Joseph K, Kannathal N, Lim CM, Suri JS. Heart rate variability: a review. Med Biol Eng Comput. 2006; 44:1031–51.

3. Elming H, Brendorp B, Kober L, Sahebzadah N, Torp-Petersen C. QTc interval in the assessment of cardiac risk. Card Electrophysiol Rev. 2002; 6:289–94.
4. Choi Y, Lim HS, Chung D, Choi JG, Yoon D. Risk evaluation of azithromycin-induced QT prolongation in real-world practice. Biomed Res Int. 2018; 2018:1574806.

5. Wilke RA, Lin DW, Roden DM, Watkins PB, Flockhart D, Zineh I, et al. Identifying genetic risk factors for serious adverse drug reactions: current progress and challenges. Nat Rev Drug Discov. 2007; 6:904–16.

6. Maheshwari A, Norby FL, Roetker NS, Soliman EZ, Koene RJ, Rooney MR, et al. Refining prediction of atrial fibrillation-related stroke using the P2-CHA2DS2-VASc score. Circulation. 2019; 139:180–91.
7. Attia ZI, Noseworthy PA, Lopez-Jimenez F, Asirvatham SJ, Deshmukh AJ, Gersh BJ, et al. An artificial intelligence-enabled ECG algorithm for the identification of patients with atrial fibrillation during sinus rhythm: a retrospective analysis of outcome prediction. Lancet. 2019; 394:861–7.

8. Attia ZI, Kapa S, Lopez-Jimenez F, McKie PM, Ladewig DJ, Satam G, et al. Screening for cardiac contractile dysfunction using an artificial intelligence-enabled electrocardiogram. Nat Med. 2019; 25:70–4.

9. van Putten M, Olbrich S, Arns M. Predicting sex from brain rhythms with deep learning. Sci Rep. 2018; 8:3069.

10. Rashkov G, Bobe A, Fastovets D, Komarova M. Natural image reconstruction from brain waves: a novel visual BCI system with native feedback. bioRxiv. 2019; 787101.

11. Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D, et al. Visual explanations from deep networks via gradient-based localization. arXiv e-prints 2016. Available from https://ui.adsabs.harvard.edu/abs/2016arXiv161002391S.

12. Rawat W, Wang Z. Deep convolutional neural networks for image classification: a comprehensive review. Neural Comput. 2017; 29:2352–449.

14. Chung J, Gulcehre C, Cho K, Bengio Y. Empirical evaluation of gated recurrent neural networks on sequence modeling. arXiv e-prints 2014. Available from https://ui.adsabs.harvard.edu/abs/2014arXiv1412.3555C.
15. Bahdanau D, Cho K, Bengio Y. Neural machine translation by jointly learning to align and translate. arXiv e-prints 2014. Available from https://arxiv.org/abs/1409.0473.
16. Liu J, Gong X. Attention mechanism enhanced LSTM with residual architecture and its application for protein-protein interaction residue pairs prediction. BMC Bioinformatics. 2019; 20:609.

17. Munkhdalai T, Liu F, Yu H. Clinical relation extraction toward drug safety surveillance using electronic health record narratives: classical learning versus deep learning. JMIR Public Health Surveill. 2018; 4:e29.

18. Galloway CD, Valys AV, Shreibati JB, Treiman DL, Petterson FL, Gundotra VP, et al. Development and validation of a deep-learning model to screen for hyperkalemia from the electrocardiogram. JAMA Cardiol. 2019; 4:428–36.

19. Yoon D, Lim HS, Jeong JC, Kim TY, Choi JG, Jang JH, et al. Quantitative evaluation of the relationship between T-wave-based features and serum potassium level in real-world clinical practice. Biomed Res Int. 2018; 2018:3054316.

20. Xia Y, Wulan N, Wang K, Zhang H. Detecting atrial fibrillation by deep convolutional neural networks. Comput Biol Med. 2018; 93:84–92.

21. Baloglu UB, Talo M, Yildirim O, Tan RS, Acharya UR. Classification of myocardial infarction with multi-lead ECG signals and deep CNN. Pattern Recognit Lett. 2019; 122:23–30.

22. Coult J, Blackwood J, Sherman L, Rea TD, Kudenchuk PJ, Kwok H. Ventricular fibrillation waveform analysis during chest compressions to predict survival from cardiac arrest. Circ Arrhythm Electrophysiol. 2019; 12:e006924.

23. Daoud H, Bayoumi M. Deep learning approach for epileptic focus localization. IEEE Trans Biomed Circuits Syst. 2019; In press.

24. Zeng H, Wu Z, Zhang J, Yang C, Zhang H, Dai G, et al. EEG emotion classification using an improved SincNet-based deep learning model. Brain Sci. 2019; 9:E326.

25. Mumtaz W, Qayyum A. A deep learning framework for automatic diagnosis of unipolar depression. Int J Med Inform. 2019; 132:103983.

26. Hannun AY, Rajpurkar P, Haghpanahi M, Tison GH, Bourn C, Turakhia MP, et al. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat Med. 2019; 25:65–9.

27. Perez MV, Mahaffey KW, Hedlin H, Rumsfeld JS, Garcia A, Ferris T, et al. Large-scale assessment of a smartwatch to identify atrial fibrillation. N Engl J Med. 2019; 381:1909–17.

28. Spartano NL, Davis-Plourde KL, Himali JJ, Andersson C, Pase MP, Maillard P, et al. Association of accelerometer-measured light-intensity physical activity with brain volume: the framingham heart study. JAMA Netw Open. 2019; 2:e192745.
29. Block VJ, Bove R, Zhao C, Garcha P, Graves J, Romeo AR, et al. Association of continuous assessment of step count by remote monitoring with disability progression among adults with multiple sclerosis. JAMA Netw Open. 2019; 2:e190570.

30. Medical device software licensing guidelines. Secondary medical device software licensing guidelines. Available from http://www.nifds.go.kr/brd/m_15/view.do?seq=12438&srchFr=&srchTo=&srchWord=&srchTp=&itm_seq_1=0&itm_seq_2=0&multi_itm_seq=0&company_cd=&company_nm=&page=4.
31. Artificial intelligence and machine learning in software as a medical device. Secondary artificial intelligence and machine learning in software as a medical device. Available from https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-software-medical-device.
32. Johnson AE, Pollard TJ, Shen L, Lehman LW, Feng M, Ghassemi M, et al. MIMIC-III, a freely accessible critical care database. Sci Data. 2016; 3:160035.

33. Pollard TJ, Johnson AEW, Raffa JD, Celi LA, Mark RG, Badawi O. The eICU collaborative research database, a freely available multi-center database for critical care research. Sci Data. 2018; 5:180178.

Fig. 1.
Potentiality and usability of hidden information present in diverse types of biosignals. The characteristic of artificial intelligence that can extract valuable information allows the opportunity to acquire and use hidden information not perceived by humans. It provides a diagnosis that is more accurate, early detection of a clinical event, and new insight on disease and prescription of drugs.
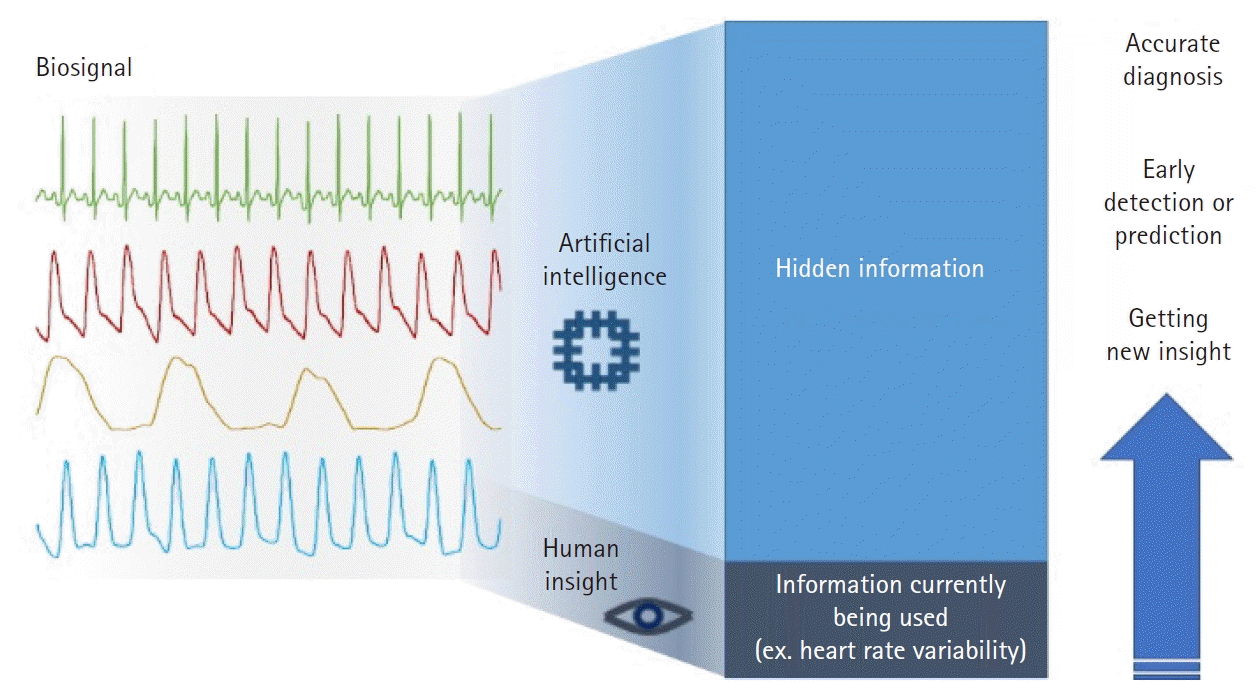
Fig. 2.
An overview of the relationship between artificial intelligence, machine learning, and deep learning. The term artificial intelligence (AI) includes all software that mimics the cognitive functions of humans. Among the various algorithms in AI, machine learning algorithms (deep learning, in particular) that learn patterns in data are considered promising owing to their distinctive performance. CNN: convolutional neural networks, RNN: recurrent neural networks, DQN: deep Q netowrk.
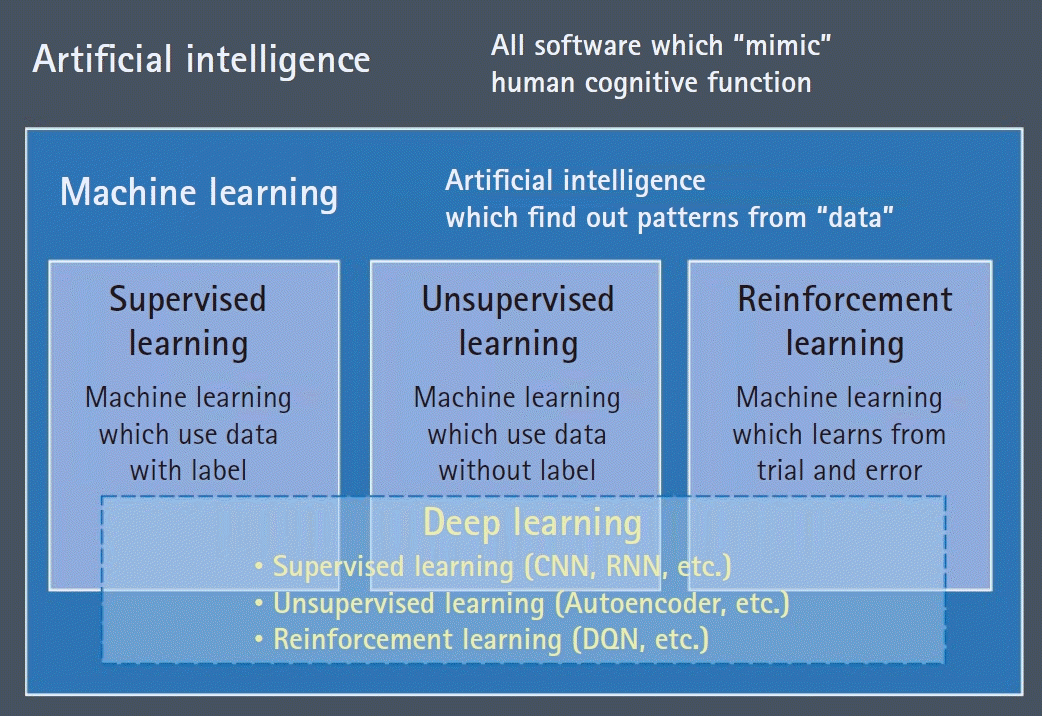
Fig. 3.
Schematic illustration of the underlying principle of convolution in a convolutional neural network (CNN) and expression of a recursive neural network (RNN). (A) Convolution is a type of matrix multiplication; however, a CNN changes the weight values in the filter in the training process to maximize the valuable features that are useful to discriminate between labels. (B) RNN recursively uses the output of its network as the input for the next step, thus, a long expression (top) is summarized as a short one (bottom).
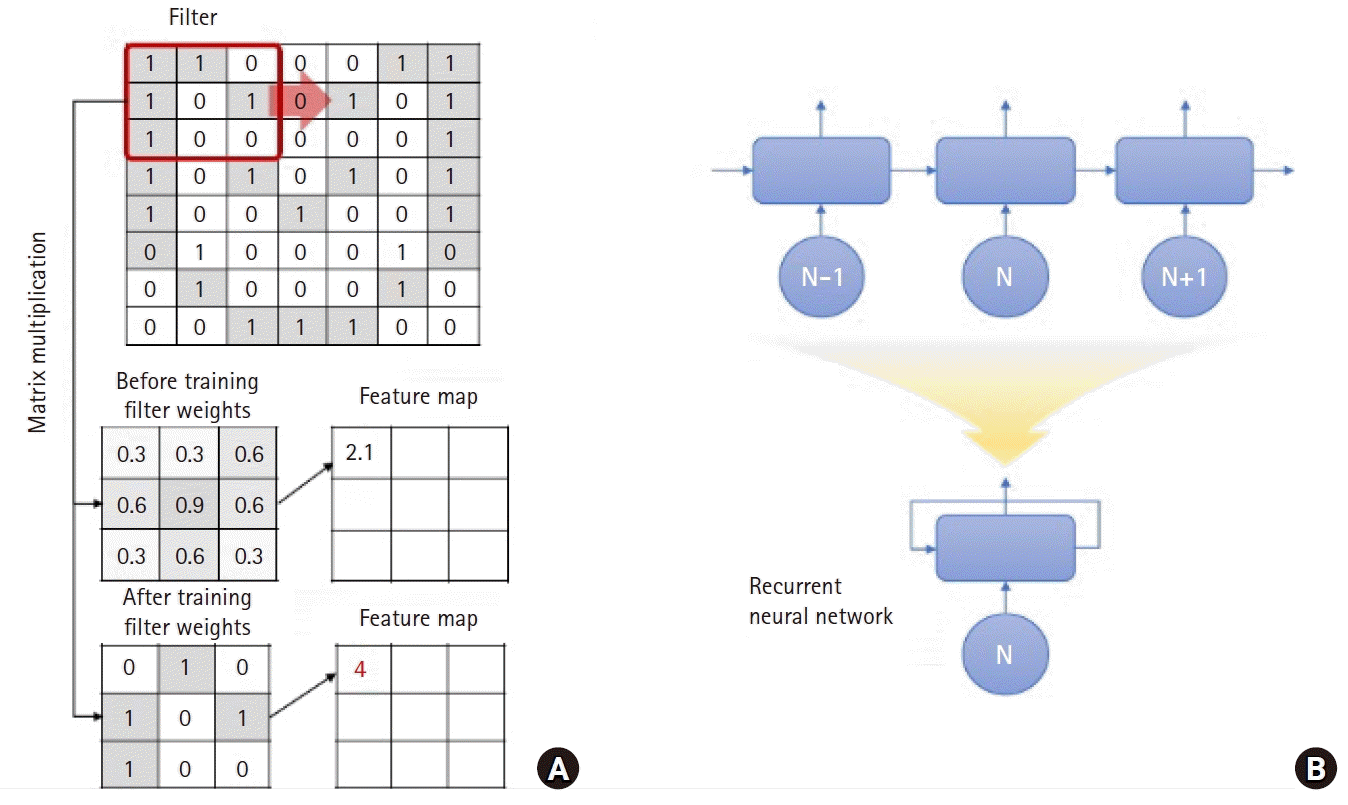
Fig. 4.
Two different approaches for extracting hidden information from biosignals in deep learning. (A) Convolution in a convolutional neural network fits the convolutional matrix to extract a feature map that is a type of matrix containing valuable information for discriminating between labels. (B) An autoencoder extracts hidden information useful for reconstruction of raw data from the data itself. Although the features are derived regardless of label information, principal information included in the raw signal is obtained that can be used in other machine learning algorithms to predict or detect labels.
Fig. 5.
Schematic of the entire process of an artificial intelligence (AI) research or project. As machine learning algorithms depend on the amount and quality of data, collecting proper datasets is a critical step to begin AI-related projects. Extracting valuable information from a huge dataset is cumbersome; hence, this step is conducted in the process of training either in a convolutional neural network or independently (Autoencoder).




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download