Abstract
Purpose
Excessive alcohol consumption has been linked to an increased risk of colorectal cancer (CRC). We evaluated the association between alcohol-related genetic variants and CRC risk.
Materials and Methods
The study cohort consisted of 5,435 CRC cases and 3,553 population-based cancer-free controls. Genotype data were generated from germline DNA using the Infinium OncoArray-500K BeadChip in 2,535 cases and 2,287 controls and the Infinium Multi-Ethnic Global BeadChip in 2,900 cases and 1,266 controls. The associations between aldehyde dehydrogenase 2 (ALDH2) rs671 and alcohol dehydrogenase 1B (ADH1B) rs1229984 polymorphisms and CRC risk were assessed using multivariate logistic regression analyses.
Results
Compared with the major homozygous ALDH2 genotype (GG), heterozygous or minor homozygous ALDH2 genotype (GA or AA, related to a low alcohol consumption) was significantly associated with a reduced risk for CRC in men (odds ratio [OR], 0.78; 95% confidence interval [CI], 0.68 to 0.90), but not in women (OR, 0.70; 95% CI, 0.47 to 1.05). A stronger association was found among regular drinkers (OR, 0.58; 95% CI, 0.47 to 0.71 in men and OR, 0.33; 95% CI, 0.18 to 0.58 in women). No association of CRC risk with ADH1B rs1229984 genotype was found. The association between alcohol-related combined genotypes and risk of CRC was significant (p for linear=0.001). The combined genotype with the highest genetically predicted alcohol consumption (ALDH2 rs671 GG and ADH1B rs1229984 AG/GG) was associated with a high risk for CRC (OR, 1.35; 95% CI, 1.11 to 1.63).
In South Korea, colorectal cancer (CRC) is the third most common cancer in men and women and the second leading cause of cancer-related death in men [1]. CRC is associated with various lifestyle factors, such as diet, physical inactivity, smoking, and alcohol consumption. Among these factors, alcohol consumption is a primary risk factor, with a linear dose–risk relation [2]. However, the strength of the association between light or moderate alcohol consumption remains controversial [3]. A recent study found no significant association between moderate alcohol intake and CRC risk [4], while a protective effect of low alcohol consumption against CRC has been reported [5,6]. These discrepancies may have resulted from the lack of uniformity in alcohol consumption measures.
Alcohol dehydrogenase 1B (ADH1B) and aldehyde dehydrogenase 2 (ALDH2) are alcohol metabolism-related genetic variants common among Eastern Asians. They have been widely used as an indicator of alcohol exposure in investigations of alcohol consumption as a risk factor for various diseases, including CRC [7]. Alcohol is metabolized to acetaldehyde and eventually acetate by ADH and ALDH, respectively. Among individuals of different ethnicities, different genetic variants in the genes encoding these enzymes can be found. Certain polymorphisms in ADH1B and ALDH2 are particularly common in East Asians, affecting drinking behaviors [8–12]. The ADH1B rs1229984 polymorphism, which is common in East Asians, decreases the activity of ADH1B. Similarly, another polymorphism common in East Asians, ALDH2 rs671, decreases the activity of ALDH2. Differences in the activity of enzymes involved in alcohol metabolism affect the levels of acetaldehyde accumulated after alcohol consumption. Notably, the A allele (major allele) of ADH1B rs1229984 and A allele (minor allele) of ALDH2 rs671 have been associated with acetaldehyde accumulation after alcohol consumption. Because acetaldehyde is responsible for the unfavorable symptoms associated with alcohol consumption, including nausea, palpitation, and headache, individuals carrying these alleles tend to avoid alcohol intake. Consequently, ALDH2 or ADH1B polymorphisms have been consistently associated with drinking behaviors [8–12]. A study that evaluated alcohol consumption in South Koreans according to ALDH2 rs671 polymorphisms showed that men with GA or AA genotypes consumed less than 12 g alcohol (approximately one drink) per day, which was considerably less than the amount consumed by men with a GG genotype [12]. Similarly, Chinese individuals carrying the GA genotype consumed less than 17 g alcohol per day [8].
Several studies have assessed the association between polymorphisms in alcohol metabolizing enzymes, including ALDH2 and ADH1B, and the risk for CRC in Eastern Asians [13–16] and Europeans [17]. However, most have been conducted on small study populations, and heterogeneity among different studies has been substantial [18,19]. Therefore, we performed a case-control study in a large Korean cohort to examine the association between particular alcohol metabolism-related genetic variants and CRC risk.
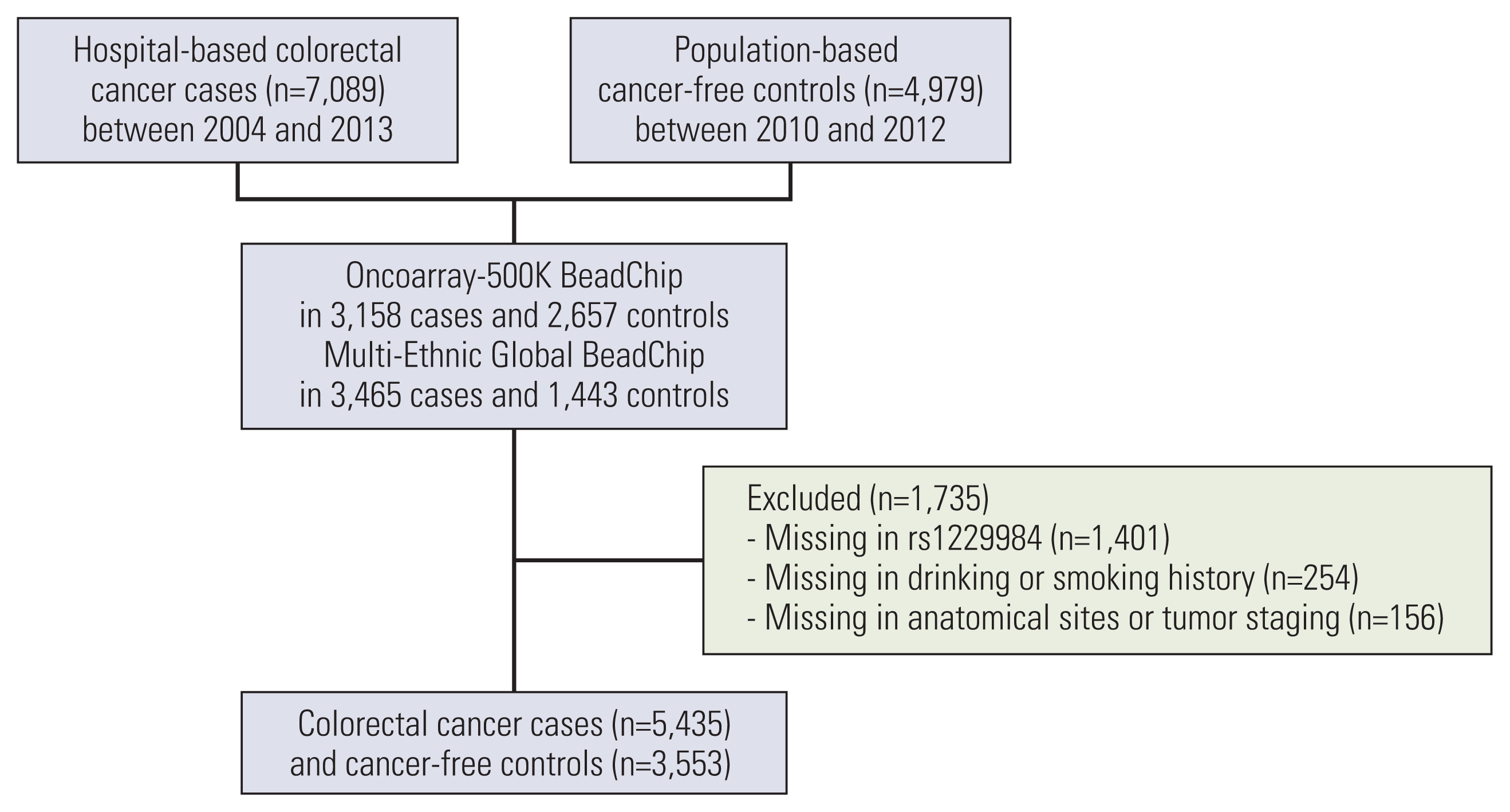
Fig. 1 shows the flow chart of this study. This study cohort consisted of 7,089 hospitalized patients and 4,979 population-based controls from the Hwasun Cancer Epidemiology Study–Colon and Rectum Cancer (HCES-CRC) [20]. In brief, the hospitalized cases were patients diagnosed with histologically confirmed CRC at Chonnam National University Hwasun Hospital between 2004 and 2013. Control individuals were recruited between 2010 and 2012 and did not develop cancer until 2017. Genome-wide association study (GWAS) analyses were performed in 6,623 cases and 4,100 controls. Of these, 866 cases and 535 controls were excluded because of missing information on rs1229984. Hence, our analyses included data from 5,435 cases and 3,553 controls without missing information (Fig. 1). CRC was classified according to the anatomical site as per the International Classification of Disease 10th edition (ICD-10) as proximal colon (C18.0–18.4), distal colon (C18.5–18.7), rectum (C19–20), and others (C18.8–18.9).
Details on genotyping and GWAS quality control were described previously [21,22]. Briefly, germline DNA genotyping was performed using the Infinium OncoArray-500K BeadChip (Oncoarray, Illumina, San Diego, CA) in 3,158 cases and 2,657 controls, and Infinium Multi-Ethnic Global BeadChip (MEGA) in 3,465 cases and 1,443 controls. For genome-wide imputation, we used the 1,000 Genome Project Phase 3 East Asian panel as a reference [23]. Imputation was conducted separately for the two genotyping arrays using IMPUTE2 ver. 2.3.2 [24]. Rs671 was genotyped, and the rs1229984 genotype was imputed in both arrays. The imputation quality of rs1229984 was sufficient for subsequent analyses (imputed information score > 0.9) [25].
Information on smoking history and alcohol consumption was obtained from the medical records of the case group and interview questionnaires administered to the control group. Individuals were classified as current drinkers or abstainers for alcohol consumption. In addition, study participants were classified as current smokers, ex-smokers, or never smokers in the medical records; we re-classified participants as ever-smokers (current/ex-smokers) or never smokers.
The baseline characteristics of the case and control participants were assessed according to sex. Comparisons in the baseline characteristics were performed using t tests for continuous variables and chi-square tests for categorical variables.
Multivariate logistic regression analyses were performed to assess the association between ALDH2 rs671/ADH1B rs1229984 polymorphisms and CRC risk. The genotypes were recategorized into major homozygotes (GG in ALDH2 rs671 or AA in ADH1B rs1229984) and other genotypes (heterozygotes and minor homozygotes). All analyses were age-adjusted. Based on the association between alcohol consumption and alcohol-related genotypes [8,10–12], we compared the CRC risk between combined genotypes in increasing genetically predicted alcohol consumption order (ALDH2 rs671 GA/AA and ADH1B rs1229984 AA, ALDH2 rs671 GA/AA and ADH1B rs1229984 AG/GG, ALDH2 rs671 GG and ADH1B rs1229984 AA, and ALDH2 rs671 GG and ADH1B rs1229984 AG/GG). Wald test was used to evaluate the interaction. p < 0.05 were considered statistically significant. All analyses were performed using R (ver. 3.6.1, R Foundation for Statistical Computing, Vienna, Austria).
The baseline characteristics of the study participants according to sex are summarized in Table 1. In both men and women, current drinkers and ever-smokers were less frequent in the case group. Women in the control group were significantly older than those in the case group, whereas the men’s age did not differ significantly between the two groups. The rectum was the most common tumor site, both in men (53.8%) and women (48.2%) (Table 1).
The distribution of ALDH2 and ADH1B polymorphisms, as well as the association between ALDH2/ADH1B genotypes and CRC risk, are listed in Tables 2 and 3. Rs671 and rs1229984 are missense mutations of the ALDH2 gene on chromosome 12 and ADH1B gene on chromosome 4, respectively. Rs671 A allele was associated with low activity of ALDH2, and its frequency was 0.147. Rs1229984 G allele was associated with high activity of ADH1B, and its frequency was 0.23. Compared to the most common genotypes, the risk for CRC was lower in men carrying the minor ALDH2 allele (A allele), but not in women. The odds ratios (ORs) for ALDH2 rs671 GA and AA genotypes were 0.78 (95% confidence interval [CI], 0.68 to 0.90) and 0.70 (95% CI, 0.47 to 1.05) in men, and 1.10 (95% CI, 0.95 to 1.27) and 1.37 (95% CI, 0.88 to 2.13) in women, respectively. We found no association between the ADH1B rs1229984 polymorphism and CRC risk in men or women. For combined genotype analyses, we used participants with ALDH2 rs671 GA/AA and ADH1B rs1229984 AA as a reference, because they had the lowest genetically predicted alcohol consumption. The risk for CRC was higher in men with genotypes associated with high alcohol consumption than in those with low alcohol consumption-related genotypes (p=0.001). The combined genotype with the highest genetically predicted alcohol consumption (ALDH2 rs671 GG and ADH1B rs1229984 AG/GG) was associated with a high risk for CRC. ORs for individuals with ALDH2 rs671 GA/AA and ADH1B rs1229984 AG/GG, ALDH2 rs671 GG and ADH1B rs1229984 AA, and ALDH2 rs671 GG and ADH1B rs1229984 AG/GG were 0.92 (95% CI, 0.73 to 1.16), 1.19 (95% CI, 0.99 to 1.42), and 1.35 (95% CI, 1.11 to 1.63), respectively. The association between alcohol-related genotypes and CRC risk was similar according to the anatomical site of CRC. In women, there was no significant association between alcohol consumption-associated genotypes and CRC risk (Table 3). However, in the subgroup analysis for anatomical sites of CRC, compared to women with ALDH2 rs671 GG genotype, those with ALDH2 rs671 AA genotype had a higher risk of distal colon cancer (OR, 1.99; 95% CI, 1.04 to 3.61).
The association between ALDH2/ADH1B genotypes and CRC risk according to drinking and smoking habits are shown in Tables 4 and 5. The impact of drinking on CRC risk was higher in women than in men. In men, the CRC risk in ALDH2 rs671 GA/AA carriers was 0.58 (95% CI, 0.47 to 0.71) for current drinkers and 0.74 (95% CI, 0.61 to 0.91) for abstainers, while in women the CRC risk in ALDH2 rs671 GA/AA carriers was 0.33 (95% CI, 0.18 to 0.58) for current drinkers and 0.88 (95% CI, 0.74 to 1.04) for abstainers. The interaction between alcohol consumption and the ALDH2 genotype was statistically significant only in women (p < 0.001). Also, the interaction of drinking status for association between combined alcohol-related genotypes and CRC was significant only in women.
Table 6 represents the association between rs671 and rs1229984 polymorphism and prevalence of current drinkers. The A allele of both rs671 and rs1229984 was related to low prevalence of current drinker in both sexes. The prevalence of current drinkers was higher in both sexes as the combined genotypes predicted to have higher alcohol consumption (p for trend < 0.001). On the other hand, the prevalence of smoker was not related to rs671 or rs1229984 polymorphism (data not shown).
We found that the association between alcohol-related genetic variants and CRC risk differed between men and women. Men with ALDH2 rs671 GA/AA had a lower risk for CRC than those with ALDH2 rs671 GG. A stronger association was found among regular drinkers than abstainer or never drinker in both sexes. However, ADH1B polymorphisms were not significantly associated with CRC in men or women.
We found that the ADH1B rs1229984 polymorphism was not significantly associated with the risk for CRC; however, heterozygous or minor homozygous ALDH2 (GA or AA) was significantly associated with a lower risk for CRC, consistent with the findings of several previous studies [13,14,16, 26]. Furthermore, our findings are largely in line with meta-analyses showing an association between ALDH2 rs671 GA/AA and low risk for CRC [18,19]. By contrast, the association between ADH1B and CRC risk remains controversial. In Japanese individuals [13,15], ADH1B rs1229984 AG/GG is associated with a high risk for CRC; however, our study and other previous studies [14,16] found no significant association between ADH1B and CRC risk. These discrepancies might be explained by differences in the effects of different single nucleotide polymorphisms on alcohol consumption behavior. Compared to ALDH2 rs671, ADH1B rs1229984 is a weaker instrumental variable for alcohol consumption. ALDH2 rs671 GG was associated with a high prevalence of alcohol dependence [9,11] or alcohol consumption [10]. Nevertheless, ADH1B polymorphisms are not significantly associated with drinking behavior [11], and their effect on alcohol consumption is weaker than that of ALDH2 polymorphisms [9,10]. Moreover, in the China Kadoorie Biobank study, the largest and most recent alcohol-related MR study [8], the effects of ADH1B polymorphisms on alcohol consumption were 2–4 times smaller than those of ALDH2 polymorphisms.
Previous studies that have evaluated the association between combined ALDH2 and ADH1B genotypes and CRC risk have provided inconsistent results. Although Yin et al. [13] reported no significant differences among combined genotypes, Matsuo et al. [15] found significant gene-gene interactions. In our study, although our study cohorts size was nearly 10 times larger than that of Matsuo et al. [15] we found no gene-gene interactions. However, we found that alcohol-related genetic variants were associated with CRC risk in men and that the risk for CRC was high in carriers of high alcohol consumption-associated combined genotypes. In women, however, these associations were not significant. Several factors could have attributed to these divergences. First, alcohol consumption habits differ significantly between men and women. The differences in alcohol consumption among different genotypes are considerably smaller in women than in men [8,10,12]. However, previous studies have not conducted sex-stratified analyses [13,15]. Second, ADH1B has pleiotropic effects on ethanol metabolism. Individuals with low ADH1B activity experience less unfavorable symptoms after alcohol intake due to the lower acetaldehyde levels [27]. Thus, ADH1B rs1229984 AG/GG carriers have higher levels of alcohol than ADH1B rs1229984 AA carriers. However, low ADH1B activity prolongs exposure to acetaldehydes produced by microbes in the gastrointestinal tract due to the low ethanol elimination rate [28]. On the other hand, salivary acetaldehyde level varies depending on the distribution of microbes in the digestive tract [29], and there are few studies on the effects of alcohol-related genetic variants on the distribution or activity of microbes, so further evaluation is necessary.
In our study, the association between alcohol-related genetic variants and CRC differed by sex. Although genetic variants favorable for alcohol consumption were associated with high risk of CRC in men, but in women there was no such association. This is because the strength of the association between genetic variants and alcohol consumption is weaker in women than in men. In our study, the amount of alcohol consumption was not investigated, but in previous MR studies of Eastern Asians [8,12], the difference in alcohol consumption according to genetic variants was smaller in women than in men. In MR analysis using weak instrumental variables, it may not be possible to exclude the effect of unmeasured confounders, one of the main objectives of MR analysis (weak instrumental bias) [30]. Interestingly, in the subgroup analysis for anatomical sites of CRC, among ALDH2 rs671 polymorphism, AA genotypes were associated with high risk of distal colon cancer in women only. ALDH2 detoxified endogenous aldehyde such as 4-hydroxy-2-nonenal and malondialdehyde produced by reactive oxygen species [31]. The hormonal effect is presumed to be involved in the carcinogenic mechanism of ALDH2 because AA genotypes had a higher risk of breast cancer in the subgroup with high estrogen receptor expression [32]. However, because few studies are evaluating hormonal mediated mechanisms, future studies are needed.
A recent meta-analysis demonstrated that the risk for CRC is lowest in individuals who consume 7 g or less of alcohol per day, and increases when consumption is 14 g/day [6]. However, we found that in men, the risk for CRC increased with increasing genetically predicted alcohol consumption. Although the China Kadoorie Biobank study [8] showed a J-shaped association between subjective alcohol consumption and the risk for cerebrovascular diseases, the association between genetically predicted alcohol consumption and cerebrovascular diseases was linear. Therefore, the protective effect of alcohol reported by studies involving questionnaires is presumed to be due to sick-quitter or under-reporter bias [8,33].
To the best of our knowledge, this is the first study on this topic involving sex-stratified analyses. In addition, we performed combined genotype analyses using a large study cohort. The results of our study using genetic variants are less affected by reverse causality or residual confounding than those of studies using subjective alcohol consumption. However, there were several limitations to this study. The mechanism of ethanol-associated carcinogenesis was not fully evaluated. The levels of accumulated acetaldehyde after alcohol intake differ between individuals with ALDH2 and ADH1B genotypes [27,28]. Therefore, to evaluate the carcinogenic effects of acetaldehyde on CRC, the dose-response relationship of alcohol should be evaluated after cohort stratification based on alcohol-related genotypes. Because we did not investigate the level of alcohol consumption, we could not evaluate such associations. In addition, the dose–response relationship between genetically predicted alcohol consumption and CRC risk could not be assessed, because the alcohol consumption based on alcohol-related genotypes was not estimated. Therefore, it is necessary to estimate the causal effect of alcohol consumption on CRC risk through the two-stage least square method in the genetic study investigating the amount of alcohol consumption, or perform a two-sample MR study using the results of GWAS on the amount of alcohol consumption.
In conclusion, the association between alcohol-related genotypes and CRC risk differed between men and women in this South Korean cohort. Men with genotypes associated with high alcohol consumption had a high risk for CRC. Future studies are required to elucidate the role of ethanol in carcinogenesis in individuals with different ethanol metabolism-related genetic variants.
Notes
Ethical Statement
This study was approved by the Institutional Review Boards (IRBs) of Chonnam National University Hospital Hwasun (CUNH IRB-2014-016). Study subjects provided their informed consent to participate the study.
Acknowledgments
This study was financially supported by Chonnam National University (grant number: 2017-2857) and US NIH (R01CA188214 and U19CA148107).
References
2. Cai S, Li Y, Ding Y, Chen K, Jin M. Alcohol drinking and the risk of colorectal cancer death: a meta-analysis. Eur J Cancer Prev. 2014; 23:532–9.
3. GBD 2016 Alcohol Collaborators. Alcohol use and burden for 195 countries and territories, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2018; 392:1015–35.
4. Klarich DS, Brasser SM, Hong MY. Moderate alcohol consumption and colorectal cancer risk. Alcohol Clin Exp Res. 2015; 39:1280–91.

5. Lee S, Woo H, Lee J, Oh JH, Kim J, Shin A. Cigarette smoking, alcohol consumption, and risk of colorectal cancer in South Korea: a case-control study. Alcohol. 2019; 76:15–21.

6. McNabb S, Harrison TA, Albanes D, Berndt SI, Brenner H, Caan BJ, et al. Meta-analysis of 16 studies of the association of alcohol with colorectal cancer. Int J Cancer. 2020; 146:861–73.

7. Cornish AJ, Tomlinson IPM, Houlston RS. Mendelian randomisation: a powerful and inexpensive method for identifying and excluding non-genetic risk factors for colorectal cancer. Mol Aspects Med. 2019; 69:41–7.

8. Millwood IY, Walters RG, Mei XW, Guo Y, Yang L, Bian Z, et al. Conventional and genetic evidence on alcohol and vascular disease aetiology: a prospective study of 500 000 men and women in China. Lancet. 2019; 393:1831–42.
9. Kim DJ, Choi IG, Park BL, Lee BC, Ham BJ, Yoon S, et al. Major genetic components underlying alcoholism in Korean population. Hum Mol Genet. 2008; 17:854–8.

10. Takeuchi F, Isono M, Nabika T, Katsuya T, Sugiyama T, Yamaguchi S, et al. Confirmation of ALDH2 as a major locus of drinking behavior and of its variants regulating multiple metabolic phenotypes in a Japanese population. Circ J. 2011; 75:911–8.

11. Quillen EE, Chen XD, Almasy L, Yang F, He H, Li X, et al. ALDH2 is associated to alcohol dependence and is the major genetic determinant of “daily maximum drinks” in a GWAS study of an isolated rural Chinese sample. Am J Med Genet B Neuropsychiatr Genet. 2014; 165B:103–10.
12. Cho Y, Kwak S, Lewis SJ, Wade KH, Relton CL, Smith GD, et al. Exploring the utility of alcohol flushing as an instrumental variable for alcohol intake in Koreans. Sci Rep. 2018; 8:458.

13. Yin G, Kono S, Toyomura K, Moore MA, Nagano J, Mizoue T, et al. Alcohol dehydrogenase and aldehyde dehydrogenase polymorphisms and colorectal cancer: the Fukuoka Colorectal Cancer Study. Cancer Sci. 2007; 98:1248–53.

14. Chiang CP, Jao SW, Lee SP, Chen PC, Chung CC, Lee SL, et al. Expression pattern, ethanol-metabolizing activities, and cellular localization of alcohol and aldehyde dehydrogenases in human large bowel: association of the functional polymorphisms of ADH and ALDH genes with hemorrhoids and colorectal cancer. Alcohol. 2012; 46:37–49.

15. Matsuo K, Wakai K, Hirose K, Ito H, Saito T, Suzuki T, et al. A gene-gene interaction between ALDH2 Glu487Lys and ADH2 His47Arg polymorphisms regarding the risk of colorectal cancer in Japan. Carcinogenesis. 2006; 27:1018–23.

16. Yang H, Zhou Y, Zhou Z, Liu J, Yuan X, Matsuo K, et al. A novel polymorphism rs1329149 of CYP2E1 and a known polymorphism rs671 of ALDH2 of alcohol metabolizing enzymes are associated with colorectal cancer in a southwestern Chinese population. Cancer Epidemiol Biomarkers Prev. 2009; 18:2522–7.

17. Ferrari P, McKay JD, Jenab M, Brennan P, Canzian F, Vogel U, et al. Alcohol dehydrogenase and aldehyde dehydrogenase gene polymorphisms, alcohol intake and the risk of colorectal cancer in the European Prospective Investigation into Cancer and Nutrition study. Eur J Clin Nutr. 2012; 66:1303–8.

18. Zhao H, Liu KJ, Lei ZD, Lei SL, Tian YQ. Meta-analysis of the aldehyde dehydrogenases-2 (ALDH2) Glu487Lys polymorphism and colorectal cancer risk. PLoS One. 2014; 9:e88656.

19. Guo XF, Wang J, Yu SJ, Song J, Ji MY, Zhang JX, et al. Meta-analysis of the ADH1B and ALDH2 polymorphisms and the risk of colorectal cancer in East Asians. Intern Med. 2013; 52:2693–9.

20. Zhang B, Jia WH, Matsuda K, Kweon SS, Matsuo K, Xiang YB, et al. Large-scale genetic study in East Asians identifies six new loci associated with colorectal cancer risk. Nat Genet. 2014; 46:533–42.

21. Lu Y, Kweon SS, Tanikawa C, Jia WH, Xiang YB, Cai Q, et al. Large-scale genome-wide association study of East Asians identifies loci associated with risk for colorectal cancer. Gastroenterology. 2019; 156:1455–66.

22. Schmit SL, Edlund CK, Schumacher FR, Gong J, Harrison TA, Huyghe JR, et al. Novel common genetic susceptibility loci for colorectal cancer. J Natl Cancer Inst. 2019; 111:146–57.
23. Genomes Project Consortium. Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015; 526:68–74.
24. Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012; 44:955–9.

25. Coleman JR, Euesden J, Patel H, Folarin AA, Newhouse S, Breen G. Quality control, imputation and analysis of genome-wide genotyping data from the Illumina HumanCoreExome microarray. Brief Funct Genomics. 2016; 15:298–304.

26. Matsuo K, Hamajima N, Hirai T, Kato T, Koike K, Inoue M, et al. Aldehyde dehydrogenase 2 (ALDH2) genotype affects rectal cancer susceptibility due to alcohol consumption. J Epidemiol. 2002; 12:70–6.

27. Peng GS, Yin SJ. Effect of the allelic variants of aldehyde dehydrogenase ALDH2*2 and alcohol dehydrogenase ADH1-B*2 on blood acetaldehyde concentrations. Hum Genomics. 2009; 3:121–7.

28. Yokoyama A, Tsutsumi E, Imazeki H, Suwa Y, Nakamura C, Yokoyama T. Contribution of the alcohol dehydrogenase-1B genotype and oral microorganisms to high salivary acetaldehyde concentrations in Japanese alcoholic men. Int J Cancer. 2007; 121:1047–54.

29. Nieminen MT, Salaspuro M. Local acetaldehyde: an essential role in alcohol-related upper gastrointestinal tract carcinogenesis. Cancers (Basel). 2018; 10:11.
30. Burgess S, Thompson SG. CRP CHD Genetics Collaboration. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. 2011; 40:755–64.

31. Zhong H, Yin H. Role of lipid peroxidation derived 4-hydroxynonenal (4-HNE) in cancer: focusing on mitochondria. Redox Biol. 2015; 4:193–9.

Table 1
General characteristics of study population according to sex
Table 2
Colorectal cancer risk according to ALDH2 and ADH1B genotypes in men
| Colorectal cancer | Proximal colon cancer | Distal colon cancer | Rectal cancer | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
||||||
| No. of controls | No. of cases | OR (95% CI) | No. of cases | OR (95% CI) | No. of cases | OR (95% CI) | No. of cases | OR (95% CI) | |
| ALDH2 rs671 | |||||||||
|
|
|||||||||
| GG | 1,015 | 2,576 | 1.00 (reference) | 440 | 1.00 (reference) | 706 | 1.00 (reference) | 1,374 | 1.00 (reference) |
|
|
|||||||||
| GA | 410 | 872 | 0.78 (0.68–0.90) | 135 | 0.73 (0.58–0.92) | 219 | 0.74 (0.61–0.90) | 460 | 0.82 (0.70–0.95) |
|
|
|||||||||
| AA | 40 | 72 | 0.70 (0.47–1.05) | 18 | 1.06 (0.58–1.86) | 18 | 0.63 (0.35–1.09) | 35 | 0.64 (0.40–1.02) |
|
|
|||||||||
| GA/AAa) | 450 | 899 | 0.77 (0.67–0.89) | 153 | 0.76 (0.61–0.95) | 237 | 0.73 (0.61–0.88) | 495 | 0.80 (0.69–0.93) |
|
|
|||||||||
| ADH1B rs1229984 | |||||||||
|
|
|||||||||
| AA | 872 | 2,033 | 1.00 (reference) | 340 | 1.00 (reference) | 551 | 1.00 (reference) | 1,098 | 1.00 (reference) |
|
|
|||||||||
| AG | 519 | 1,279 | 1.08 (0.95–1.23) | 234 | 1.16 (0.94–1.41) | 333 | 1.03 (0.86–1.23) | 687 | 1.06 (0.92–1.23) |
|
|
|||||||||
| GG | 74 | 163 | 0.97 (0.73–1.31) | 19 | 0.68 (0.39–1.13) | 59 | 1.32 (0.91–1.89) | 84 | 0.90 (0.65–1.26) |
|
|
|||||||||
| AG/GGa) | 593 | 1,442 | 1.06 (0.94–1.21) | 253 | 1.10 (0.90–1.34) | 392 | 1.06 (0.90–1.26) | 771 | 1.04 (0.90–1.20) |
|
|
|||||||||
| ALDH2 rs671 & ADH1B rs1229984 | |||||||||
|
|
|||||||||
| ALDH2 GA/AA & ADH1B AA | 256 | 531 | 1.00 (reference) | 81 | 1.00 (reference) | 137 | 1.00 (reference) | 302 | 1.00 (reference) |
|
|
|||||||||
| ALDH2 GA/AA & ADH1B AG/GG | 194 | 368 | 0.92 (0.73–1.16) | 72 | 1.12 (0.77–1.62) | 100 | 0.95 (0.68–1.30) | 193 | 0.84 (0.64–1.09) |
|
|
|||||||||
| ALDH2 GG & ADH1B AA | 616 | 1,502 | 1.19 (0.99–1.42) | 259 | 1.32 (0.99–1.78) | 414 | 1.27 (1.00–1.63) | 796 | 1.10 (0.90–1.35) |
|
|
|||||||||
| ALDH2 GG & ADH1B AG/GG | 399 | 1,074 | 1.35 (1.11–1.63) | 181 | 1.47 (1.08–2.01) | 292 | 1.43 (1.11–1.86) | 578 | 1.25 (1.01–1.55) |
|
|
|||||||||
| p for linear trend | 0.001 | 0.009 | 0.001 | 0.006 | |||||
Table 3
Colorectal cancer risk according to ALDH2 and ADH1B genotypes in women
| Colorectal cancer | Proximal colon cancer | Distal colon cancer | Rectal cancer | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
||||||
| No. of controls | No. of cases | OR (95% CI) | No. of cases | OR (95% CI) | No. of cases | OR (95% CI) | No. of cases | OR (95% CI) | |
| ALDH2 rs671 | |||||||||
|
|
|||||||||
| GG | 1,541 | 1,417 | 1.00 (reference) | 376 | 1.00 (reference) | 325 | 1.00 (reference) | 701 | 1.00 (reference) |
|
|
|||||||||
| GA | 508 | 496 | 1.10 (0.95–1.27) | 137 | 1.15 (0.92–1.44) | 125 | 1.22 (0.97–1.54) | 226 | 1.02 (0.84–1.22) |
|
|
|||||||||
| AA | 39 | 47 | 1.37 (0.88–2.13) | 14 | 1.51 (0.77–2.80) | 15 | 1.99 (1.04–3.61) | 17 | 0.97 (0.53–1.72) |
|
|
|||||||||
| GA/AAa) | 547 | 543 | 1.12 (0.97–1.29) | 151 | 1.18 (0.94–1.46) | 140 | 1.27 (1.02–1.59) | 243 | 1.01 (0.85–1.21) |
|
|
|||||||||
| ADH1B rs1229984 | |||||||||
|
|
|||||||||
| AA | 1,235 | 1,153 | 1.00 (reference) | 327 | 1.00 (reference) | 256 | 1.00 (reference) | 558 | 1.00 (reference) |
|
|
|||||||||
| AG | 729 | 705 | 1.07 (0.94–1.23) | 173 | 0.92 (0.75–1.14) | 183 | 1.22 (0.98–1.51) | 338 | 1.07 (0.90–1.26) |
|
|
|||||||||
| GG | 124 | 102 | 0.99 (0.75–1.31) | 27 | 0.95 (0.60–1.45) | 26 | 1.11 (0.70–1.72) | 48 | 0.97 (0.67–1.37) |
|
|
|||||||||
| AG/GGa) | 853 | 807 | 1.06 (0.93–1.21) | 200 | 0.92 (0.76–1.13) | 209 | 1.20 (0.98–1.48) | 386 | 1.05 (0.90–1.23) |
|
|
|||||||||
| ALDH2 rs671 & ADH1B rs1229984 | |||||||||
|
|
|||||||||
| ALDH2 GA/AA & ADH1B AA | 337 | 314 | 1.00 (reference) | 94 | 1.00 (reference) | 73 | 1.00 (reference) | 144 | 1.00 (reference) |
|
|
|||||||||
| ALDH2 GA/AA & ADH1B AG/GG | 210 | 229 | 1.25 (0.98–1.60) | 57 | 1.05 (0.71–1.53) | 67 | 1.55 (1.06–2.27) | 99 | 1.19 (0.87–1.63) |
|
|
|||||||||
| ALDH2 GG & ADH1B AA | 898 | 839 | 0.98 (0.81–1.18) | 233 | 0.91 (0.69–1.20) | 183 | 0.91 (0.68–1.24) | 414 | 1.05 (0.84–1.33) |
|
|
|||||||||
| ALDH2 GG & ADH1B AG/GG | 643 | 578 | 0.98 (0.81–1.19) | 143 | 0.81 (0.60–1.09) | 142 | 0.99 (0.73–1.37) | 287 | 1.06 (0.83–1.36) |
|
|
|||||||||
| p for linear trend | 0.343 | 0.105 | 0.285 | 0.876 | |||||
Table 4
Impact of ALDH2/ADH1B genotype on CRC risk stratified by the alcohol drinking and smoking status in men
Table 5
Impact of ALDH2/ADH1B genotype on CRC risk stratified by the alcohol drinking and smoking status in women
Table 6
Associations between rs671 and rs122984 and prevalence of current drinker in control groups




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download