1. Zhu X, Albertsen PC, Andriole GL, Roobol MJ, Schroder FH, Vickers AJ. Risk-based prostate cancer screening. Eur Urol. 2012; 61:652–61.

2. Roobol MJ, Steyerberg EW, Kranse R, Wolters T, van den Bergh RC, Bangma CH, et al. A risk-based strategy improves prostate-specific antigen-driven detection of prostate cancer. Eur Urol. 2010; 57:79–85.

3. Mehralivand S, Shih JH, Rais-Bahrami S, Oto A, Bednarova S, Nix JW, et al. A magnetic resonance imaging-based prediction model for prostate biopsy risk stratification. JAMA Oncol. 2018; 4:678–85.

4. Palsdottir T, Nordstrom T, Aly M, Jaderling F, Clements M, Gronberg H, et al. A unified prostate cancer risk prediction model combining the Stockholm3 test and magnetic resonance imaging. Eur Urol Oncol. 2019; 2:490–6.

5. Cucchiara V, Cooperberg MR, Dall’Era M, Lin DW, Montorsi F, Schalken JA, et al. Genomic markers in prostate cancer decision making. Eur Urol. 2018; 73:572–82.

6. Nordstrom T, Akre O, Aly M, Gronberg H, Eklund M. Prostate-specific antigen (PSA) density in the diagnostic algorithm of prostate cancer. Prostate Cancer Prostatic Dis. 2018; 21:57–63.

7. Gadzinski AJ, Cooperberg MR. Prostate cancer markers. Cancer Treat Res. 2018; 175:55–86.

8. Ahmed HU, El-Shater Bosaily A, Brown LC, Gabe R, Kaplan R, Parmar MK, et al. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): a paired validating confirmatory study. Lancet. 2017; 389:815–22.

9. van der Leest M, Cornel E, Israel B, Hendriks R, Padhani AR, Hoogenboom M, et al. Head-to-head comparison of transrectal ultrasound-guided prostate biopsy versus multiparametric prostate resonance imaging with subsequent magnetic resonance-guided biopsy in biopsy-naive men with elevated prostate-specific antigen: a large prospective multicenter clinical study. Eur Urol. 2019; 75:570–8.
10. Radtke JP, Wiesenfarth M, Kesch C, Freitag MT, Alt CD, Celik K, et al. Combined clinical parameters and multiparametric magnetic resonance Imaging for advanced risk modeling of prostate cancer-patient-tailored risk stratification can reduce unnecessary biopsies. Eur Urol. 2017; 72:888–96.

11. van Leeuwen PJ, Hayen A, Thompson JE, Moses D, Shnier R, Bohm M, et al. A multiparametric magnetic resonance imaging-based risk model to determine the risk of significant prostate cancer prior to biopsy. BJU Int. 2017; 120:774–81.

12. Alberts AR, Roobol MJ, Verbeek JF, Schoots IG, Chiu PK, Osses DF, et al. Prediction of high-grade prostate cancer following multiparametric magnetic resonance imaging: improving the Rotterdam European Randomized Study of screening for prostate cancer risk calculators. Eur Urol. 2019; 75:310–8.

13. Porter KK, King A, Galgano SJ, Sherrer RL, Gordetsky JB, Rais-Bahrami S. Financial implications of biparametric prostate MRI. Prostate Cancer Prostatic Dis. 2020; 23:88–93.

14. Jambor I, Bostrom PJ, Taimen P, Syvanen K, Kahkonen E, Kallajoki M, et al. Novel biparametric MRI and targeted biopsy improves risk stratification in men with a clinical suspicion of prostate cancer (IMPROD Trial). J Magn Reson Imaging. 2017; 46:1089–95.

15. Kuhl CK, Bruhn R, Kramer N, Nebelung S, Heidenreich A, Schrading S. Abbreviated biparametric prostate MR imaging in men with elevated prostate-specific antigen. Radiology. 2017; 285:493–505.

16. Boesen L, Norgaard N, Logager V, Balslev I, Bisbjerg R, Thestrup KC, et al. Assessment of the diagnostic accuracy of biparametric magnetic resonance imaging for prostate cancer in biopsy-naive men: the Biparametric MRI for Detection of Prostate Cancer (BIDOC) study. JAMA Netw Open. 2018; 1:e180219.
17. Rais-Bahrami S, Siddiqui MM, Vourganti S, Turkbey B, Rastinehad AR, Stamatakis L, et al. Diagnostic value of biparametric magnetic resonance imaging (MRI) as an adjunct to prostate-specific antigen (PSA)-based detection of prostate cancer in men without prior biopsies. BJU Int. 2015; 115:381–8.

18. Sherrer RL, Glaser ZA, Gordetsky JB, Nix JW, Porter KK, Rais-Bahrami S. Comparison of biparametric MRI to full multiparametric MRI for detection of clinically significant prostate cancer. Prostate Cancer Prostatic Dis. 2019; 22:331–6.

19. Noh TI, Tae JH, Kim HK, Shim JS, Kang SG, Sung DJ, et al. Diagnostic accuracy and value of magnetic resonance imaging-ultrasound fusion transperineal targeted and template systematic prostate biopsy based on bi-parametric magnetic resonance imaging. Cancer Res Treat. 2020; 52:714–21.

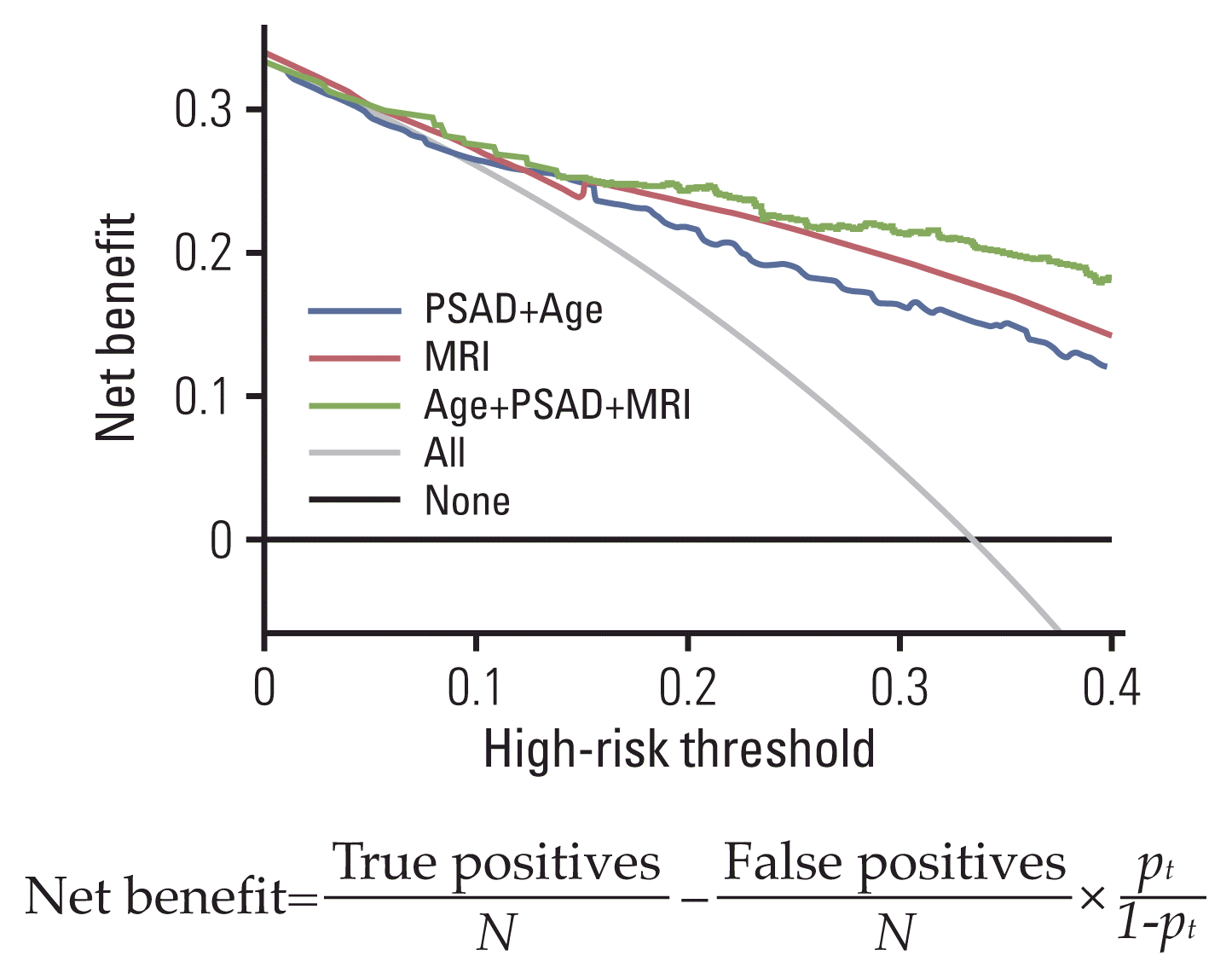
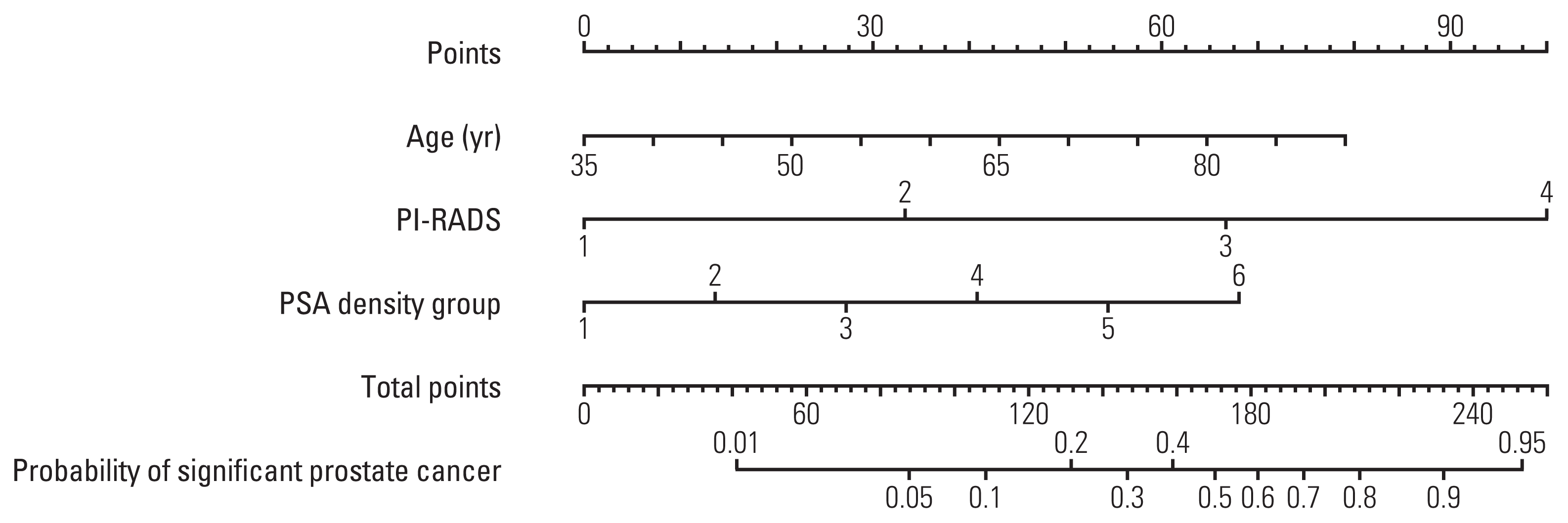
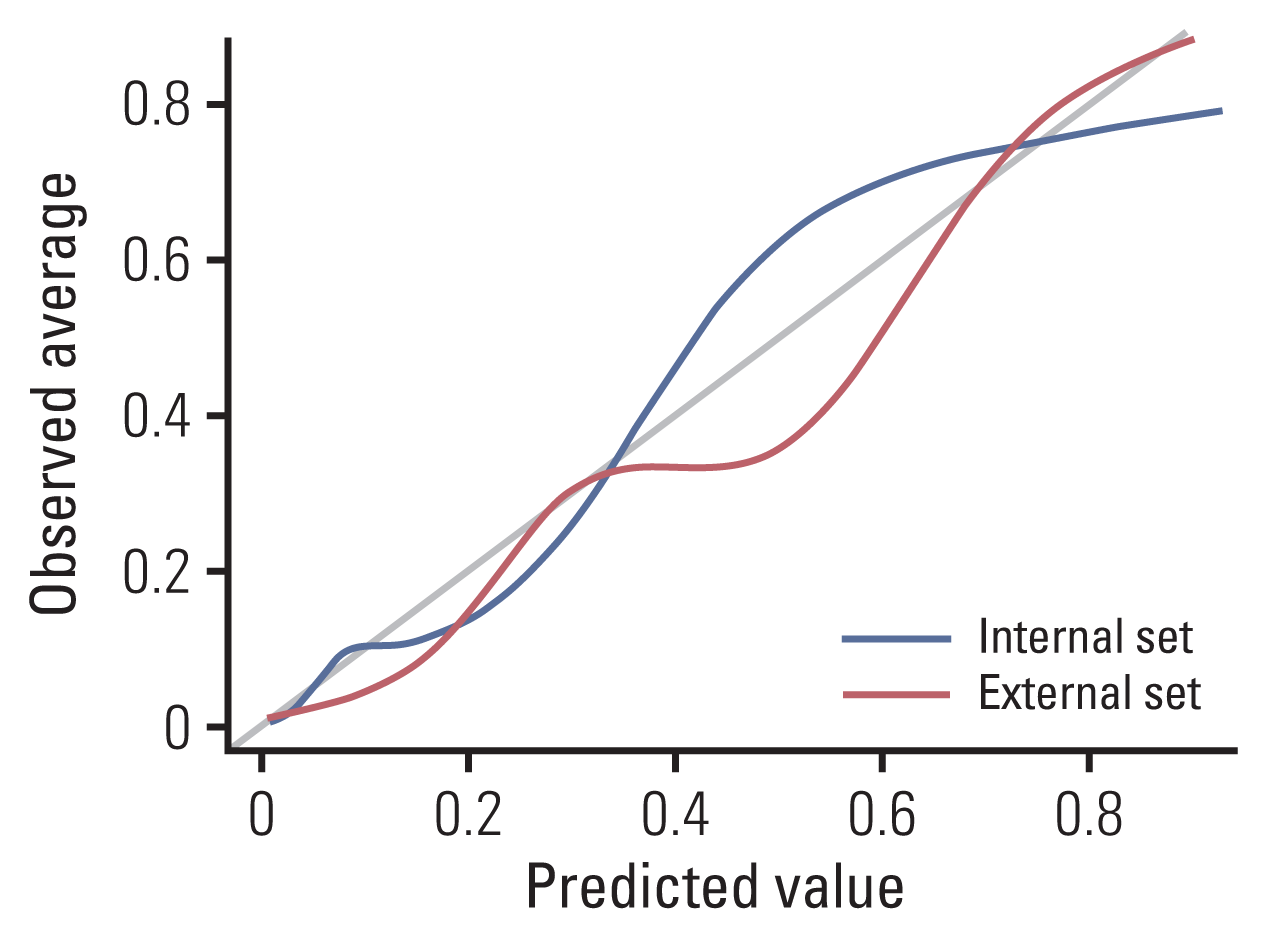
20. Vickers AJ, Van Calster B, Steyerberg EW. Net benefit approaches to the evaluation of prediction models, molecular markers, and diagnostic tests. BMJ. 2016; 352:i6.

21. de Rooij M, Israel B, Tummers M, Ahmed HU, Barrett T, Giganti F, et al. ESUR/ESUI consensus statements on multi-parametric MRI for the detection of clinically significant prostate cancer: quality requirements for image acquisition, interpretation and radiologists’ training. Eur Radiol. 2020; 30:5404–16.

22. Lee R, Localio AR, Armstrong K, Malkowicz SB, Schwartz JS. Free PSA Study Group. A meta-analysis of the performance characteristics of the free prostate-specific antigen test. Urology. 2006; 67:762–8.

23. Mikolajczyk SD, Marks LS, Partin AW, Rittenhouse HG. Free prostate-specific antigen in serum is becoming more complex. Urology. 2002; 59:797–802.

24. Pereira-Azevedo N, Verbeek JF, Nieboer D, Bangma CH, Roobol MJ. Head-to-head comparison of prostate cancer risk calculators predicting biopsy outcome. Transl Androl Urol. 2018; 7:18–26.

25. Kretschmer A, Tilki D. Biomarkers in prostate cancer: current clinical utility and future perspectives. Crit Rev Oncol Hematol. 2017; 120:180–93.
26. Osses DF, Roobol MJ, Schoots IG. Prediction medicine: biomarkers, risk calculators and magnetic resonance imaging as risk stratification tools in prostate cancer diagnosis. Int J Mol Sci. 2019; 20:1637.

27. Govers TM, Hessels D, Vlaeminck-Guillem V, Schmitz-Drager BJ, Stief CG, Martinez-Ballesteros C, et al. Cost-effectiveness of SelectMDx for prostate cancer in four European countries: a comparative modeling study. Prostate Cancer Prostatic Dis. 2019; 22:101–9.

28. Pullen L, Radtke JP, Wiesenfarth M, Roobol MJ, Verbeek JF, Wetter A, et al. External validation of novel magnetic resonance imaging-based models for prostate cancer prediction. BJU Int. 2020; 125:407–16.





 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download