1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018; 68:394–424.

3. Hsu YM, Zhang Y, You Y, Wang D, Li H, Duramad O, et al. The adaptor protein CARD9 is required for innate immune responses to intracellular pathogens. Nat Immunol. 2007; 8:198–205.

4. Yang H, Minamishima YA, Yan Q, Schlisio S, Ebert BL, Zhang X, et al. pVHL acts as an adaptor to promote the inhibitory phosphorylation of the NF-kappaB agonist Card9 by CK2. Mol Cell. 2007; 28:15–27.
5. Cao Z, Conway KL, Heath RJ, Rush JS, Leshchiner ES, Ramirez-Ortiz ZG, et al. Ubiquitin ligase TRIM62 regulates CARD9-mediated anti-fungal immunity and intestinal inflammation. Immunity. 2015; 43:715–26.

6. Nakamura S, Nakamura S, Matsumoto T, Yada S, Hirahashi M, Suekane H, et al. Overexpression of caspase recruitment domain (CARD) membrane-associated guanylate kinase 1 (CARMA1) and CARD9 in primary gastric B-cell lymphoma. Cancer. 2005; 104:1885–93.

7. Leo VI, Tan SH, Bergmann H, Cheah PY, Chew MH, Lim KH, et al. CARD9 promotes sex-biased colon tumors in the APCmin mouse model. Cancer Immunol Res. 2015; 3:721–6.

8. Tan W, Hildebrandt MA, Pu X, Huang M, Lin J, Matin SF, et al. Role of inflammatory related gene expression in clear cell renal cell carcinoma development and clinical outcomes. J Urol. 2011; 186:2071–7.

9. Li H, Tang Z, Zhu H, Ge H, Cui S, Jiang W. Proteomic study of benign and malignant pleural effusion. J Cancer Res Clin Oncol. 2016; 142:1191–200.

10. Qu J, Liu L, Xu Q, Ren J, Xu Z, Dou H, et al. CARD9 prevents lung cancer development by suppressing the expansion of myeloid-derived suppressor cells and IDO production. Int J Cancer. 2019; 145:2225–37.

11. Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017; 45:D362–8.

12. Cai J, Xia L, Li J, Ni S, Song H, Wu X. Tumor-associated macrophages derived TGF-beta induced epithelial to mesenchymal transition in colorectal cancer cells through Smad2, 3-4/Snail signaling pathway. Cancer Res Treat. 2019; 51:252–66.
13. Hara H, Ishihara C, Takeuchi A, Imanishi T, Xue L, Morris SW, et al. The adaptor protein CARD9 is essential for the activation of myeloid cells through ITAM-associated and Toll-like receptors. Nat Immunol. 2007; 8:619–29.

14. Saijo S, Ikeda S, Yamabe K, Kakuta S, Ishigame H, Akitsu A, et al. Dectin-2 recognition of alpha-mannans and induction of Th17 cell differentiation is essential for host defense against Candida albicans. Immunity. 2010; 32:681–91.
15. Robinson MJ, Osorio F, Rosas M, Freitas RP, Schweighoffer E, Gross O, et al. Dectin-2 is a Syk-coupled pattern recognition receptor crucial for Th17 responses to fungal infection. J Exp Med. 2009; 206:2037–51.

16. Yang M, Shao JH, Miao YJ, Cui W, Qi YF, Han JH, et al. Tumor cell-activated CARD9 signaling contributes to metastasis-associated macrophage polarization. Cell Death Differ. 2014; 21:1290–302.

17. Bergmann H, Roth S, Pechloff K, Kiss EA, Kuhn S, Heikenwalder M, et al. Card9-dependent IL-1beta regulates IL-22 production from group 3 innate lymphoid cells and promotes colitis-associated cancer. Eur J Immunol. 2017; 47:1342–53.
18. Liu M, Luo F, Ding C, Albeituni S, Hu X, Ma Y, et al. Dectin-1 activation by a natural product beta-glucan converts immunosuppressive macrophages into an M1-like phenotype. J Immunol. 2015; 195:5055–65.
19. Haas T, Heidegger S, Wintges A, Bscheider M, Bek S, Fischer JC, et al. Card9 controls Dectin-1-induced T-cell cytotoxicity and tumor growth in mice. Eur J Immunol. 2017; 47:872–9.

20. Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010; 141:39–51.

21. An J, Liu H, Magyar CE, Guo Y, Veena MS, Srivatsan ES, et al. Hyperactivated JNK is a therapeutic target in pVHL-deficient renal cell carcinoma. Cancer Res. 2013; 73:1374–85.

22. LeibundGut-Landmann S, Gross O, Robinson MJ, Osorio F, Slack EC, Tsoni SV, et al. Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat Immunol. 2007; 8:630–8.

23. Pereira M, Tourlomousis P, Wright J, Monie TP, Bryant CE. CARD9 negatively regulates NLRP3-induced IL-1beta production on Salmonella infection of macrophages. Nat Commun. 2016; 7:12874.

24. Chen Z, Liu F, Zhang N, Cao D, Liu M, Tan Y, et al. p38beta, A novel regulatory target of Pokemon in hepatic cells. Int J Mol Sci. 2013; 14:13511–24.
25. Tomas-Loba A, Manieri E, Gonzalez-Teran B, Mora A, Leiva-Vega L, Santamans AM, et al. p38gamma is essential for cell cycle progression and liver tumorigenesis. Nature. 2019; 568:557–60.
26. Malik A, Sharma D, Malireddi RK, Guy CS, Chang TC, Olsen SR, et al. SYK-CARD9 signaling axis promotes gut fungimediated inflammasome activation to restrict colitis and colon cancer. Immunity. 2018; 49:515–30.

27. Wang T, Fan C, Yao A, Xu X, Zheng G, You Y, et al. The adaptor protein CARD9 protects against colon cancer by restricting mycobiota-mediated expansion of myeloid-derived suppressor cells. Immunity. 2018; 49:504–14.

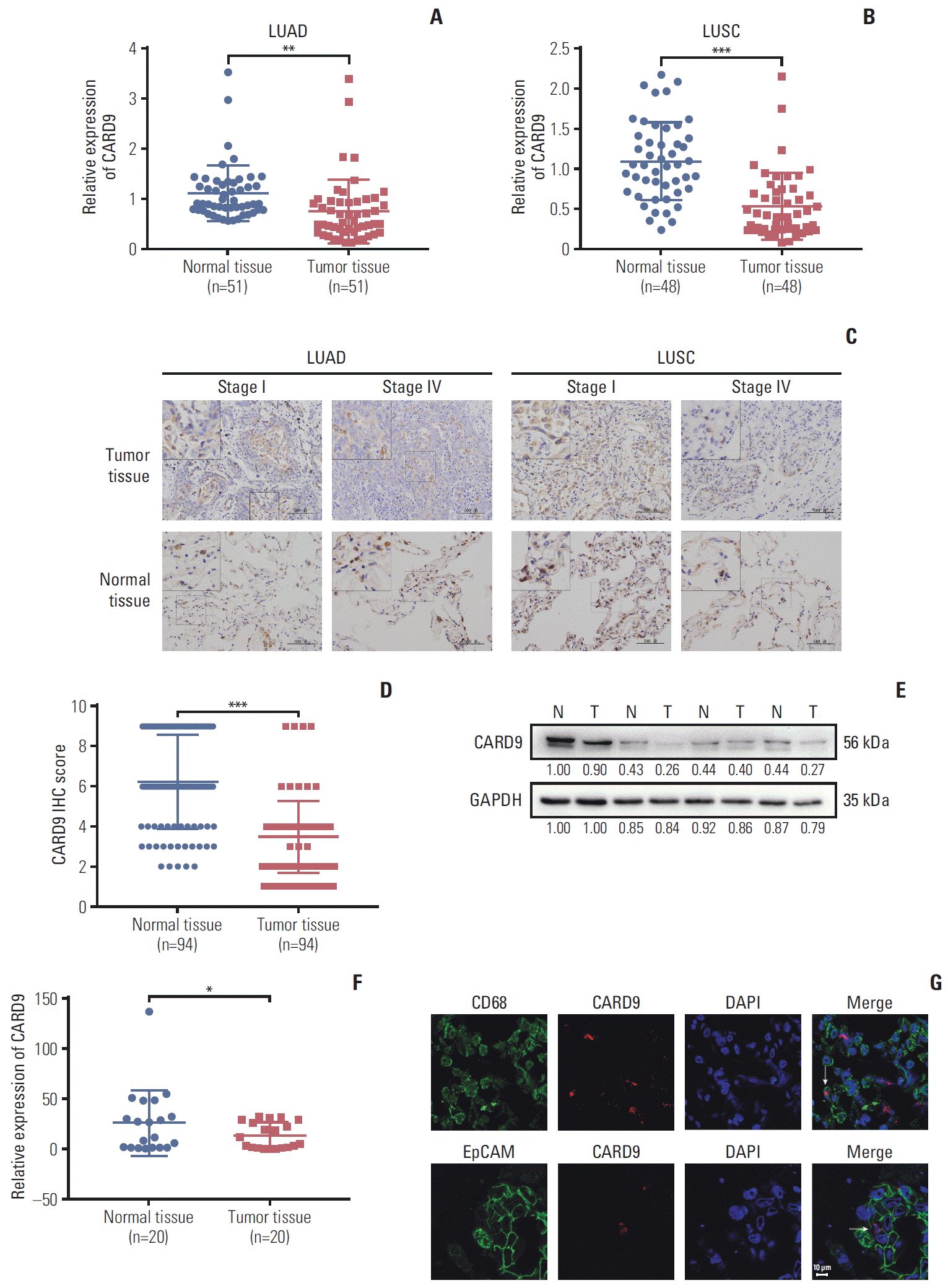
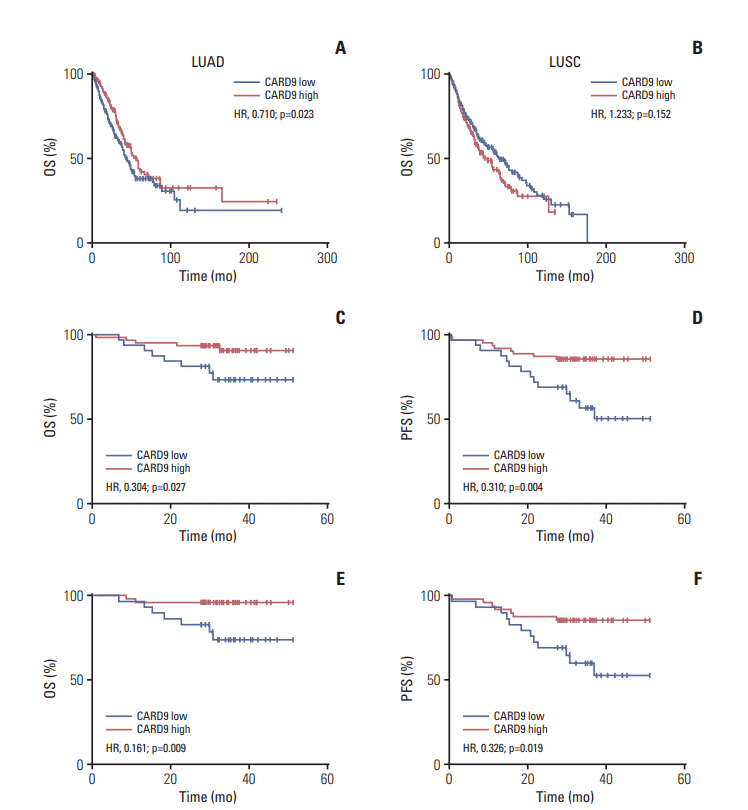
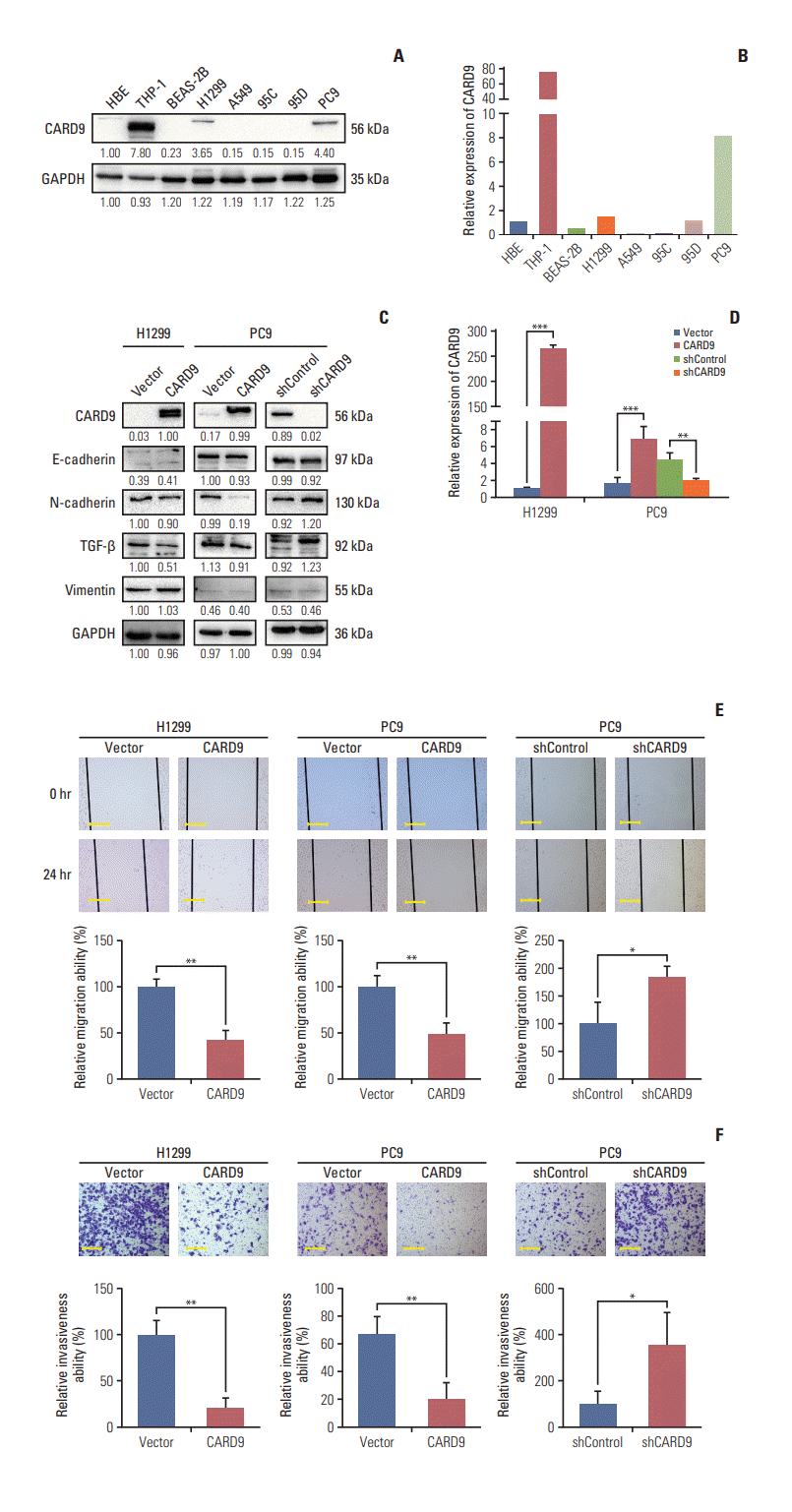
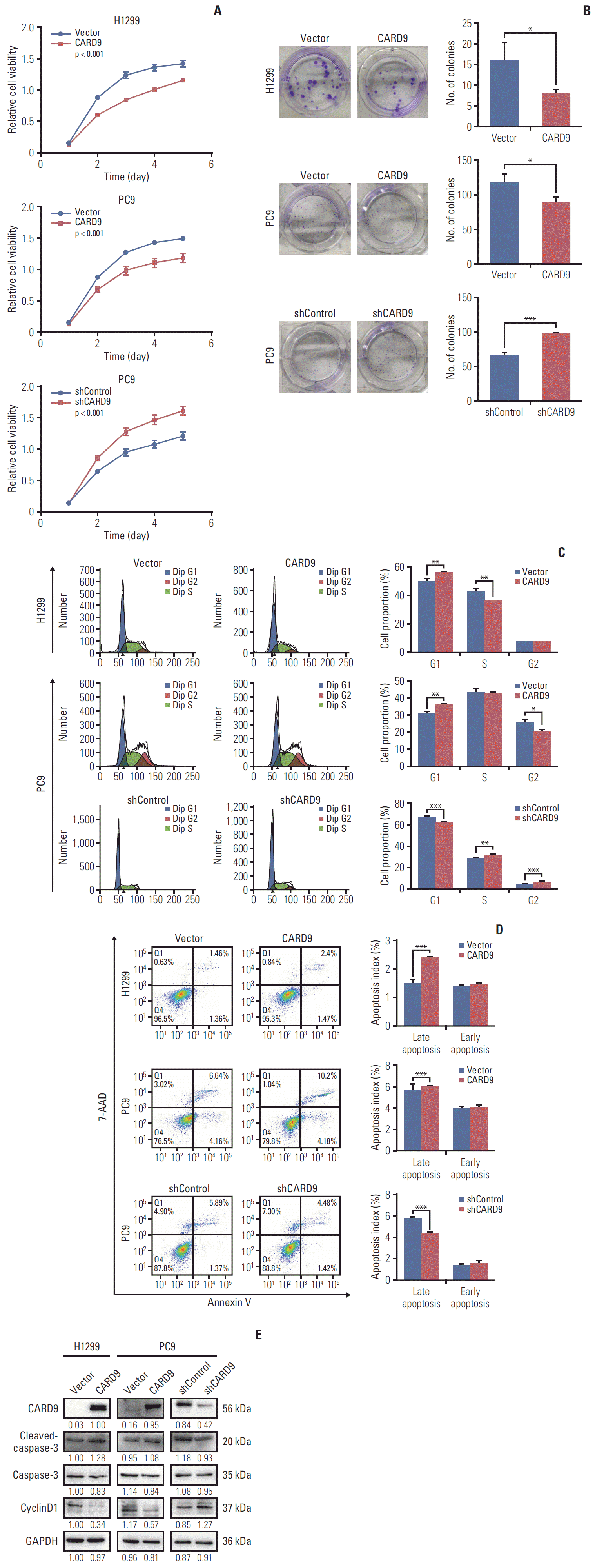




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download