1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017; 67:7–30.

2. Jiang X, Tang H, Chen T. Epidemiology of gynecologic cancers in China. J Gynecol Oncol. 2018; 29:e7.

3. Oronsky B, Ray CM, Spira AI, Trepel JB, Carter CA, Cottrill HM. A brief review of the management of platinum-resistant-platinum-refractory ovarian cancer. Med Oncol. 2017; 34:103.

4. Nikpayam E, Tasharrofi B, Sarrafzadeh S, Ghafouri-Fard S. The role of long non-coding RNAs in ovarian cancer. Iran Biomed J. 2017; 21:3–15.

5. Vergote I, Trope CG, Amant F, Kristensen GB, Ehlen T, Johnson N, et al. Neoadjuvant chemotherapy or primary surgery in stage IIIC or IV ovarian cancer. N Engl J Med. 2010; 363:943–53.

6. Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014; 15:7–21.

7. Chen J, Zhang K, Song H, Wang R, Chu X, Chen L. Long noncoding RNA CCAT1 acts as an oncogene and promotes chemoresistance in docetaxel-resistant lung adenocarcinoma cells. Oncotarget. 2016; 7:62474–89.

8. Nissan A, Stojadinovic A, Mitrani-Rosenbaum S, Halle D, Grinbaum R, Roistacher M, et al. Colon cancer associated transcript-1: a novel RNA expressed in malignant and pre-malignant human tissues. Int J Cancer. 2012; 130:1598–606.

9. Deng L, Yang SB, Xu FF, Zhang JH. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J Exp Clin Cancer Res. 2015; 34:18.

10. Ma MZ, Chu BF, Zhang Y, Weng MZ, Qin YY, Gong W, et al. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p. Cell Death Dis. 2015; 6:e1583.

11. Liu SP, Yang JX, Cao DY, Shen K. Identification of differentially expressed long non-coding RNAs in human ovarian cancer cells with different metastatic potentials. Cancer Biol Med. 2013; 10:138–41.
12. Wang Q, Zhang W, Hao S. LncRNA CCAT1 modulates the sensitivity of paclitaxel in nasopharynx cancers cells via miR-181a/CPEB2 axis. Cell Cycle. 2017; 16:795–801.

13. Yang X, Zheng F, Xing H, Gao Q, Wei W, Lu Y, et al. Resistance to chemotherapy-induced apoptosis via decreased caspase-3 activity and overexpression of antiapoptotic proteins in ovarian cancer. J Cancer Res Clin Oncol. 2004; 130:423–8.

14. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011; 146:353–8.

15. Wang S, Huang X, Lee CK, Liu B. Elevated expression of erbB3 confers paclitaxel resistance in erbB2-overexpressing breast cancer cells via upregulation of Survivin. Oncogene. 2010; 29:4225–36.

16. Zhong Y, Gao D, He S, Shuai C, Peng S. Dysregulated expression of long noncoding RNAs in ovarian cancer. Int J Gynecol Cancer. 2016; 26:1564–70.

17. Lai XJ, Cheng HF. LncRNA colon cancer-associated transcript 1 (CCAT1) promotes proliferation and metastasis of ovarian cancer via miR-1290. Eur Rev Med Pharmacol Sci. 2018; 22:322–8.
18. Inoue S, Salah-Eldin AE, Omoteyama K. Apoptosis and anticancer drug resistance. Hum Cell. 2001; 14:211–21.
19. Helm CW, States JC. Enhancing the efficacy of cisplatin in ovarian cancer treatment - could arsenic have a role. J Ovarian Res. 2009; 2:2.

20. Fraser M, Leung B, Jahani-Asl A, Yan X, Thompson WE, Tsang BK. Chemoresistance in human ovarian cancer: the role of apoptotic regulators. Reprod Biol Endocrinol. 2003; 1:66.
21. Hu B, Zhang H, Wang Z, Zhang F, Wei H, Li L. LncRNA CCAT1/miR-130a-3p axis increases cisplatin resistance in non-small-cell lung cancer cell line by targeting SOX4. Cancer Biol Ther. 2017; 18:974–83.

22. Liang HL, Hu AP, Li SL, Xie JP, Ma QZ, Liu JY. MiR-454 prompts cell proliferation of human colorectal cancer cells by repressing CYLD expression. Asian Pac J Cancer Prev. 2015; 16:2397–402.

23. Zhou L, Qu YM, Zhao XM, Yue ZD. Involvement of miR-454 overexpression in the poor prognosis of hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2016; 20:825–9.
24. Li Q, Liu J, Meng X, Pang R, Li J. MicroRNA-454 may function as an oncogene via targeting AKT in triple negative breast cancer. J Biol Res (Thessalon). 2017; 24:10.

25. Wang X, Liu B, Wen F, Song Y. MicroRNA-454 inhibits the malignant biological behaviours of gastric cancer cells by directly targeting mitogen-activated protein kinase 1. Oncol Rep. 2018; 39:1494–504.

26. Zhao X, Li X, Zhou L, Ni J, Yan W, Ma R, et al. LncRNA HOXA11-AS drives cisplatin resistance of human LUAD cells via modulating miR-454-3p/Stat3. Cancer Sci. 2018; 109:3068–79.

27. Chen X, Duan N, Zhang C, Zhang W. Survivin and tumorigenesis: molecular mechanisms and therapeutic strategies. J Cancer. 2016; 7:314–23.

28. Xu Y, Wang S, Chan HF, Lu H, Lin Z, He C, et al. Dihydromyricetin induces apoptosis and reverses drug resistance in ovarian cancer cells by p53-mediated downregulation of survivin. Sci Rep. 2017; 7:46060.

29. Chen Q, Zhang H. Smac combined with DDP can inhibit drug resistance of ovarian cancer through regulation of Survivin expression. Cancer Biomark. 2018; 22:1–6.

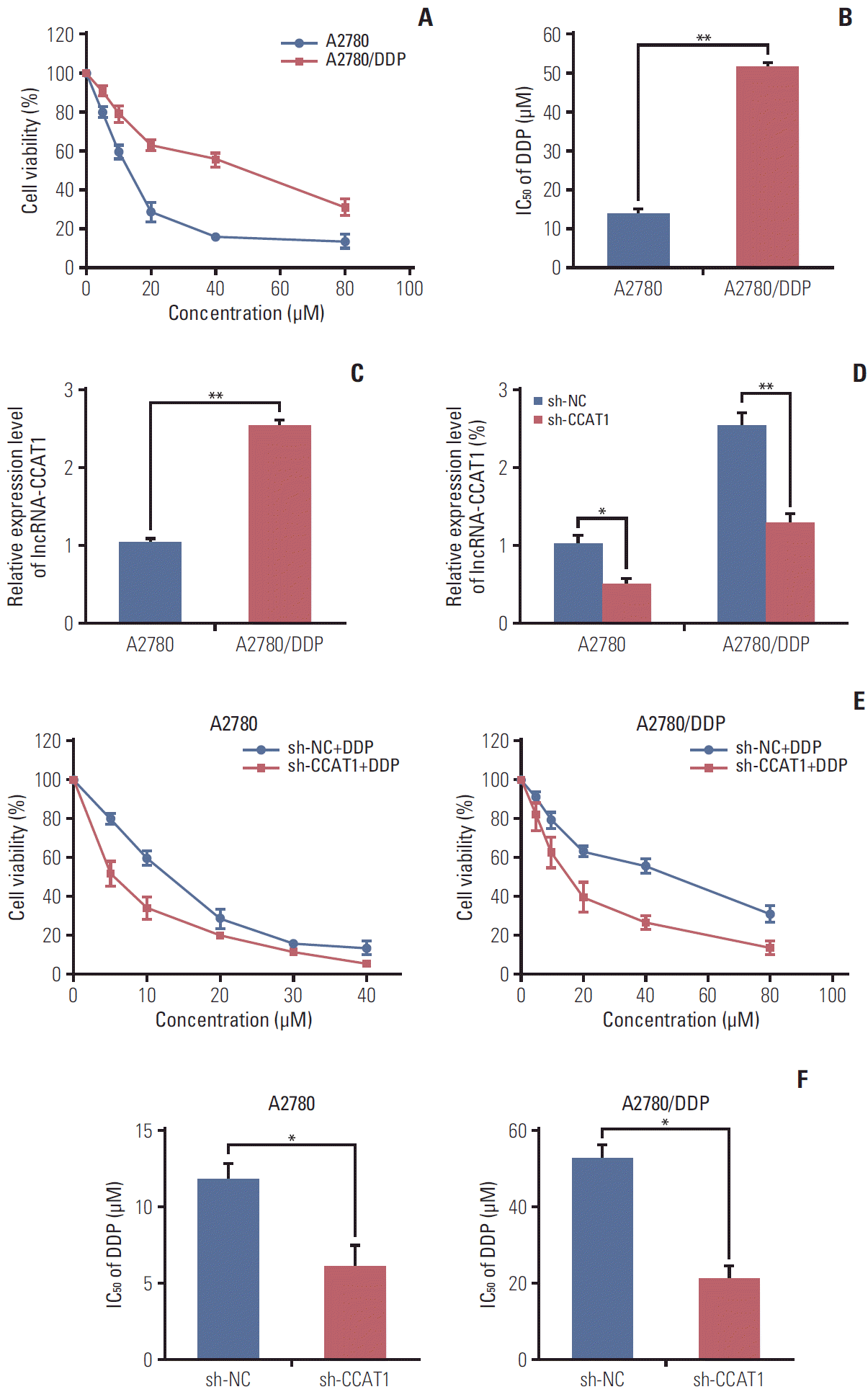
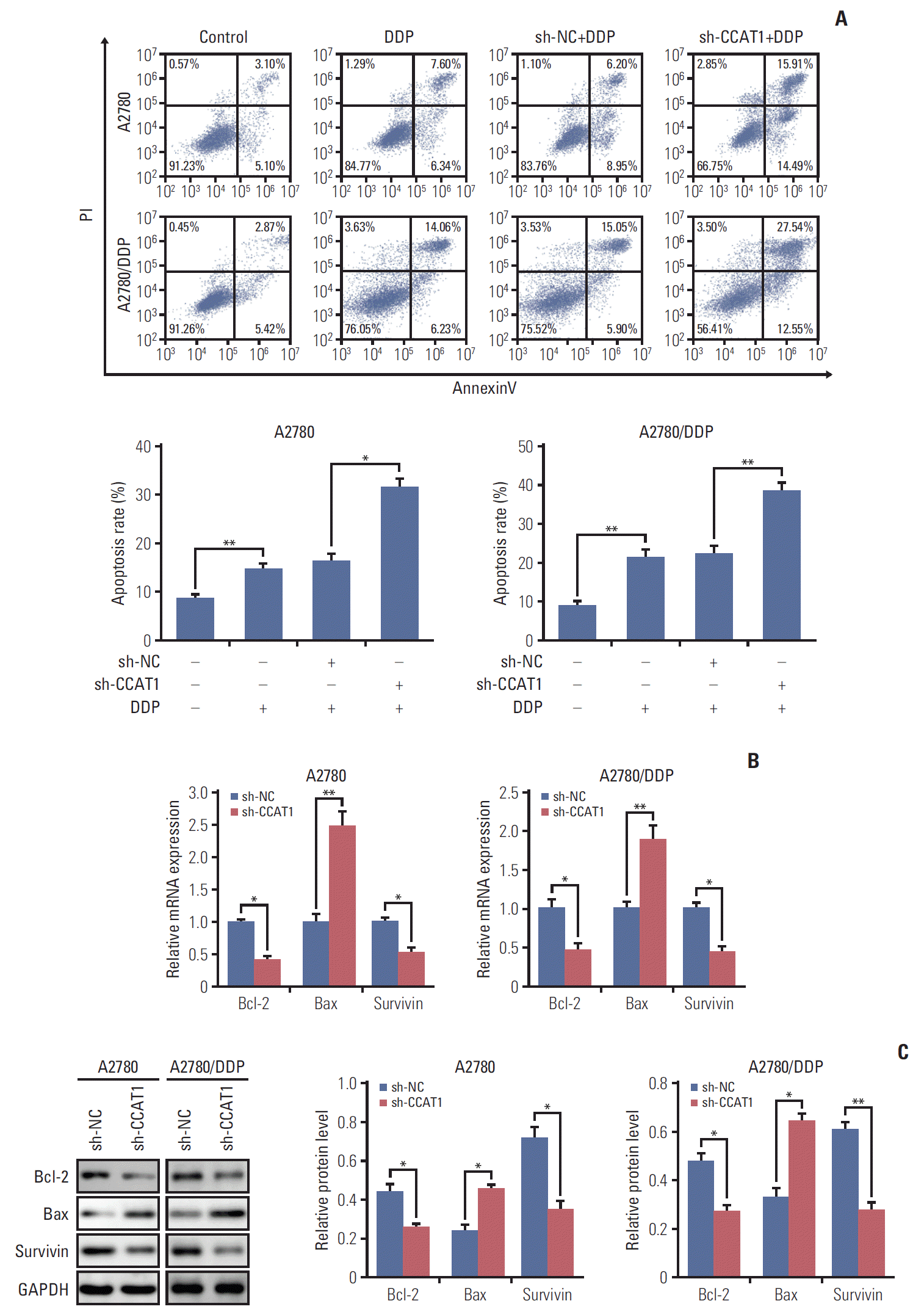
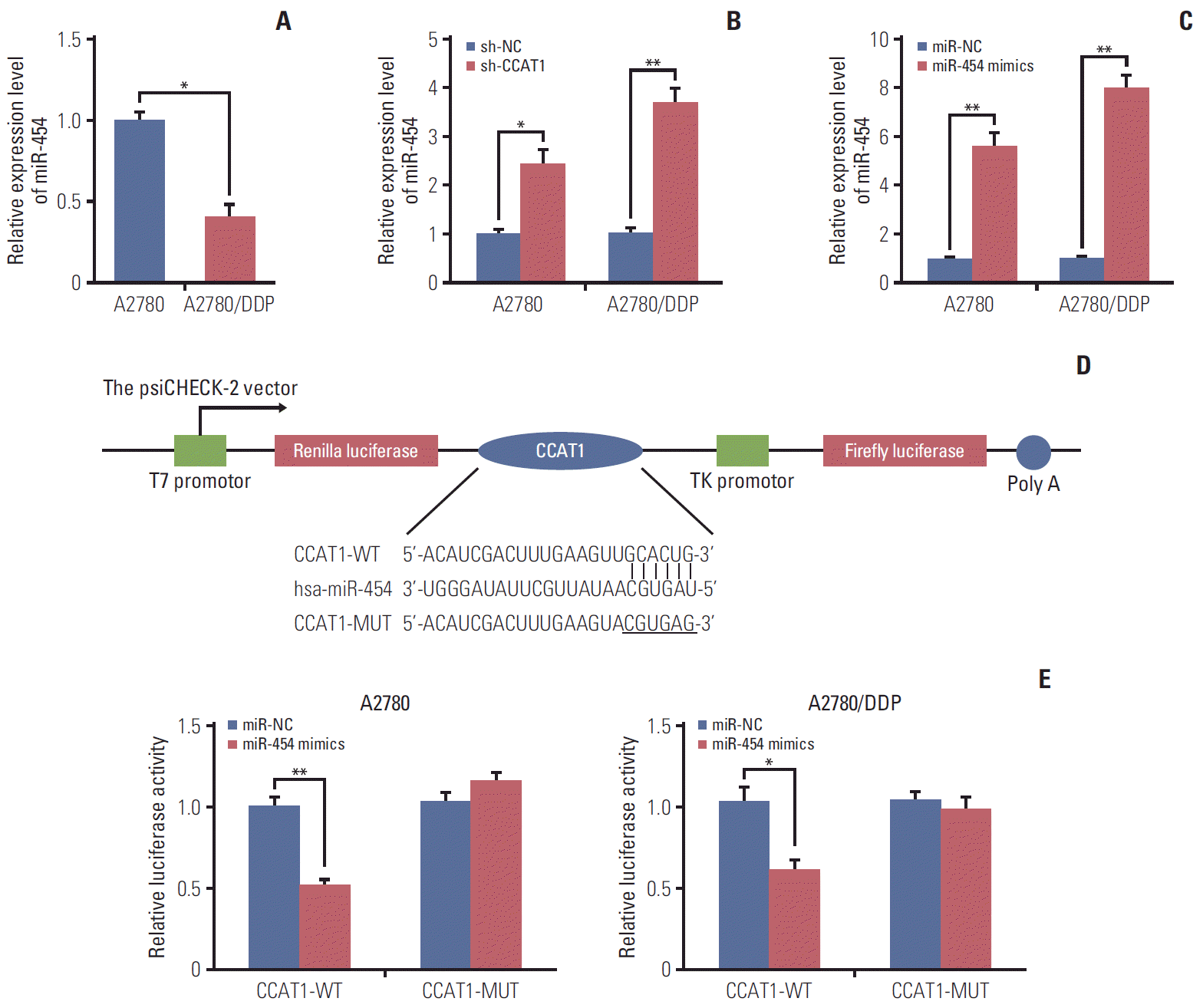
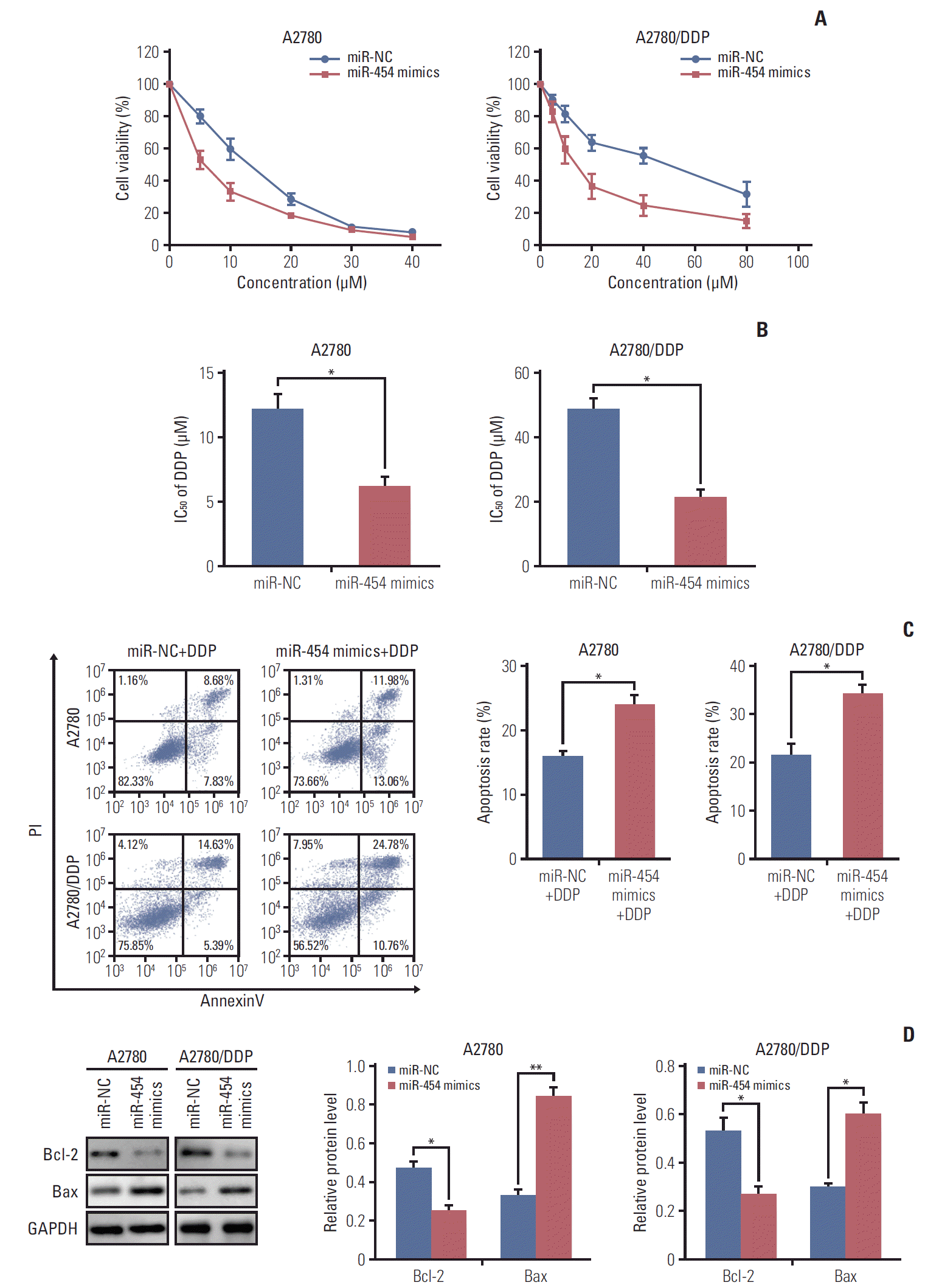
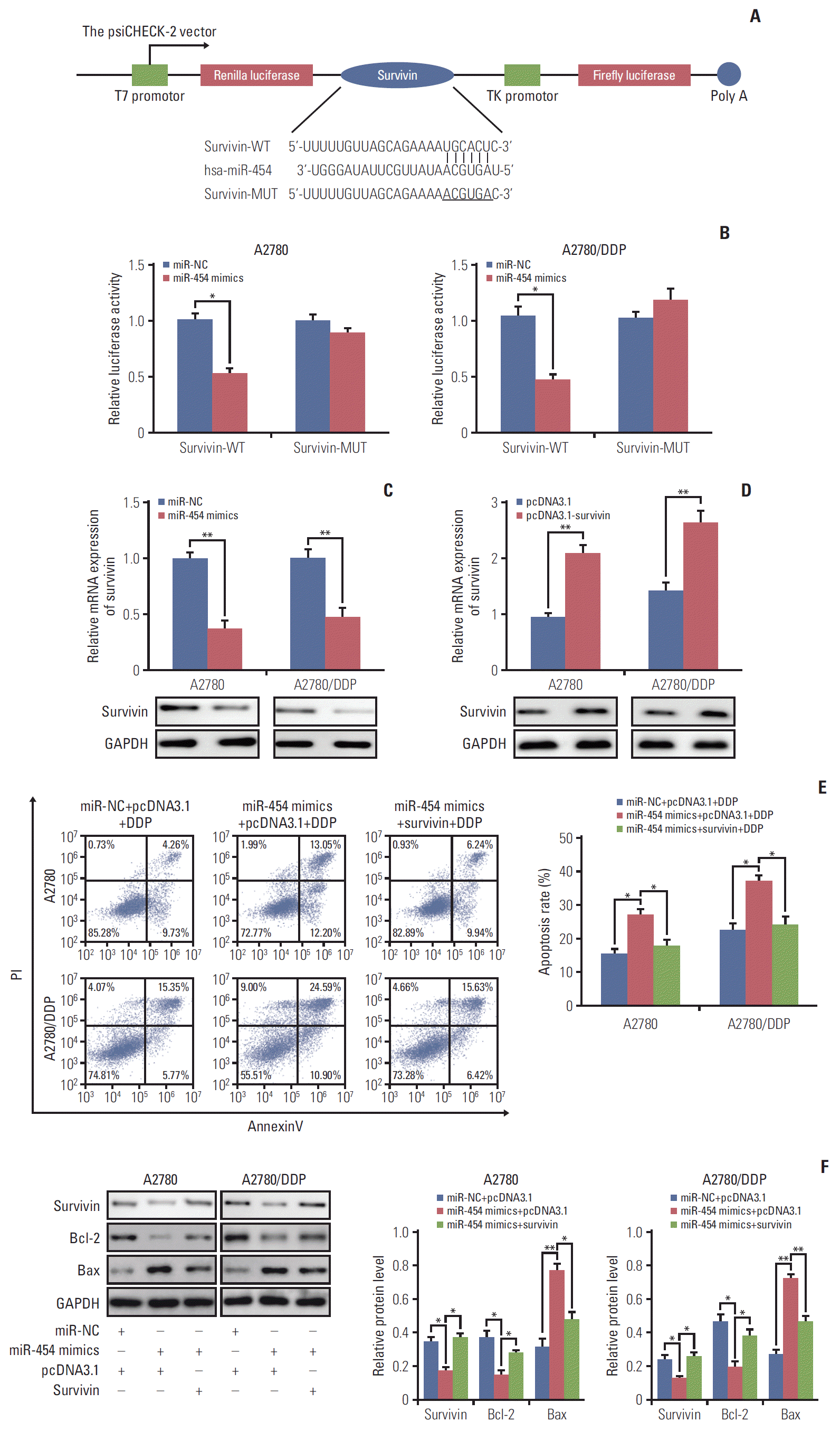
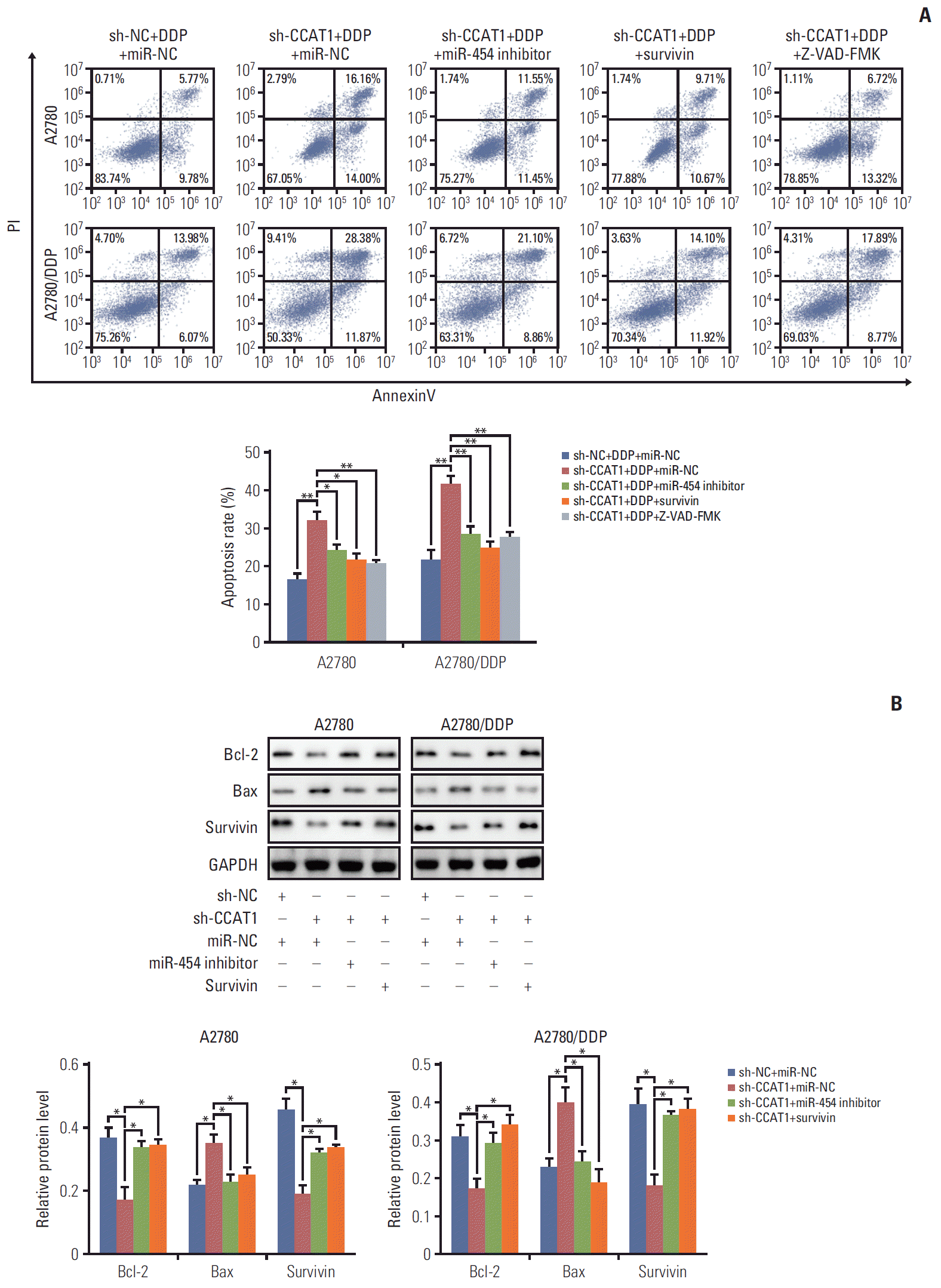
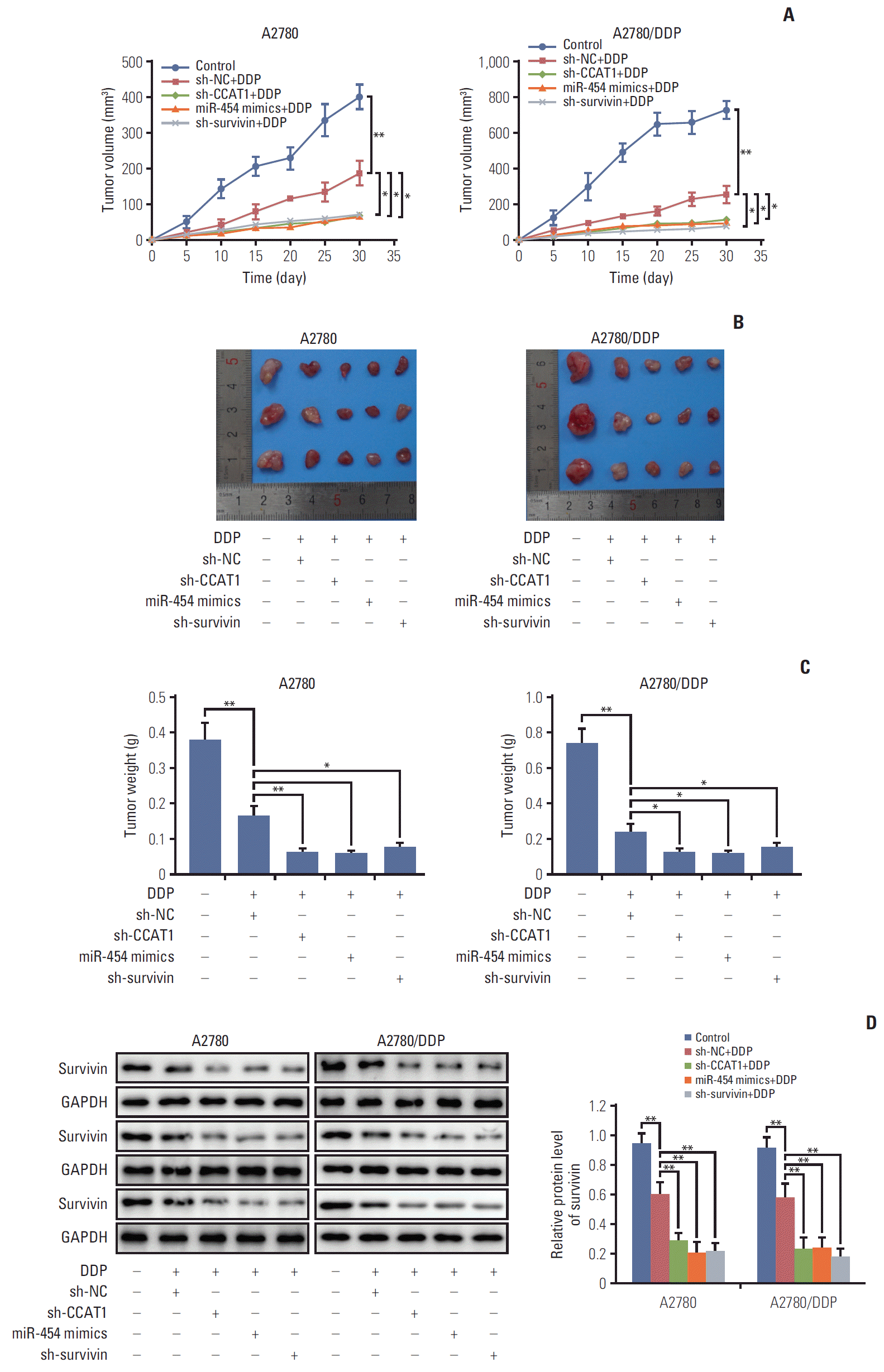




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download