Introduction
Materials and Methods
1. Cell lines and cell culture
2. Patients and tissue microarrays
3. Immunohistochemistry and scoring
4. Interference of CTSC expression in HCC cell lines
5. Quantitative real-time PCR
6. Western blot assay
7. Cell Counting Kit-8 assay
8. Colony formation
9. Animals and treatment
10. Cell migration and invasion assays
11. The bioinformatic web tools
12. Statistical analysis
13. Ethical statement
Results
1. CTSC is frequently upregulated in HCC and correlated with poor prognosis of HCC patients
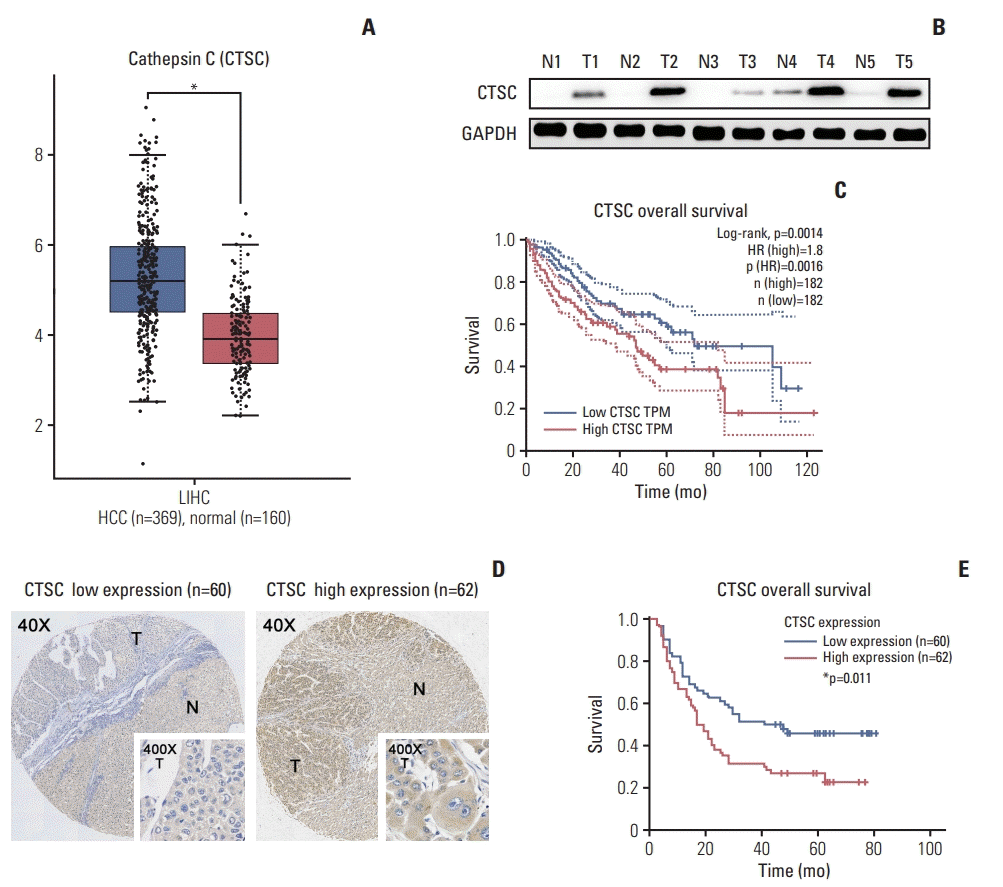 | Fig. 1.Cathepsin C (CTSC) is frequently upregulated in hepatocellular carcinoma (HCC) and correlated with poor prognosis of HCC patients. (A) The mRNA level of CTSC in HCC specimens analyzed by GEPIA web tool. (B) Western blot assay detected CTSC expression in both HCC and normal tissues. GAPDH, glyceraldehyde 3-phosphate dehydrogenase. (C) The correlation between CTSC expression and overall survival (OS) analyzed by GEPIA. (D) The differential expression in HCC specimens upon tissue microarrays. T, tumor; N, adjacent normal liver. (E) Kaplan-Meier survival analysis of OS in 122 HCC patients upon tissue microarrays. |
Table 1.
Table 2.
2. CTSC promotes HCC cells proliferation, migration, and invasion
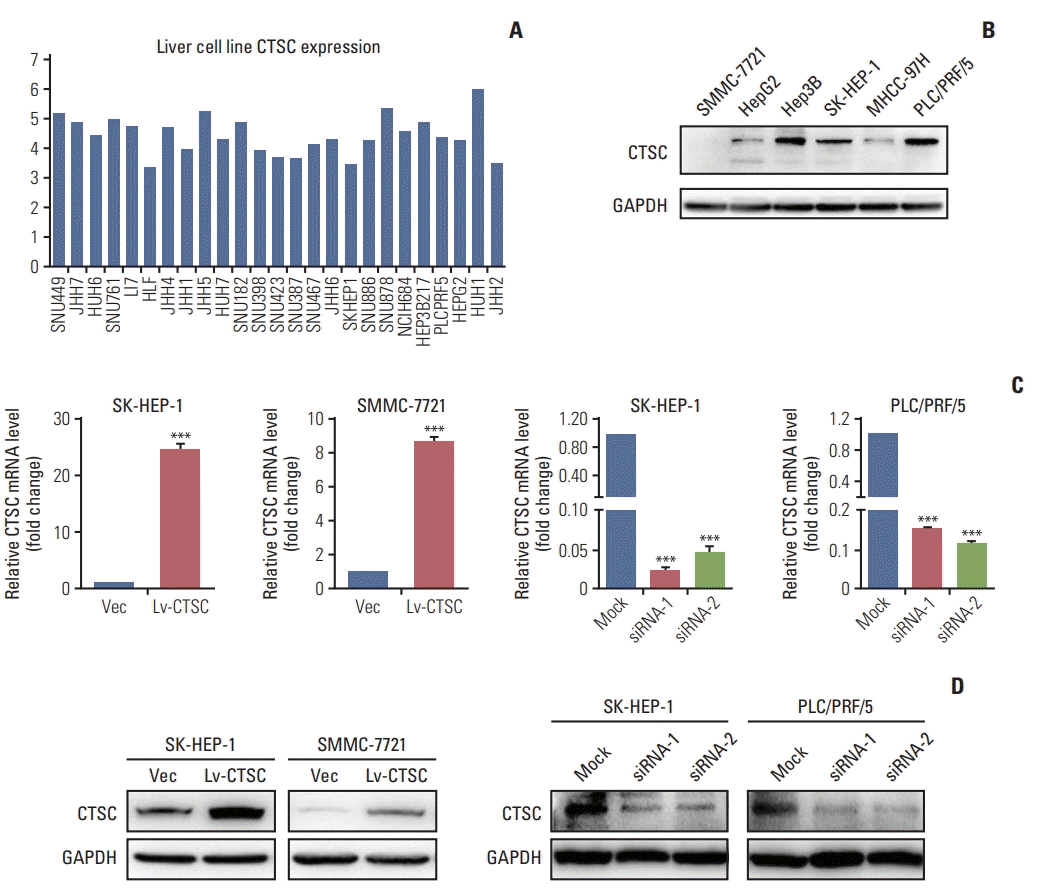 | Fig. 2.The expression of cathepsin C (CTSC) in hepatocellular carcinoma (HCC) cell lines. (A) The mRNA level in HCC cell lines analyzed by Cancer Cell Line Encyclopedia. (B) Western blot assay verified CTSC expression in HCC cell lines. GAPDH, glyceraldehyde 3-phosphate dehydrogenase. (C) The confirmation of CTSC overexpression in SH-HEP-1 and SMMC-7721 cells by quantitative real-time polymerase chain reaction. (D) Knockdown of CTSC in SK-HEP-1 and PLC/PRF/5 cells which was validated by western blot assay. ***p < 0.001. |
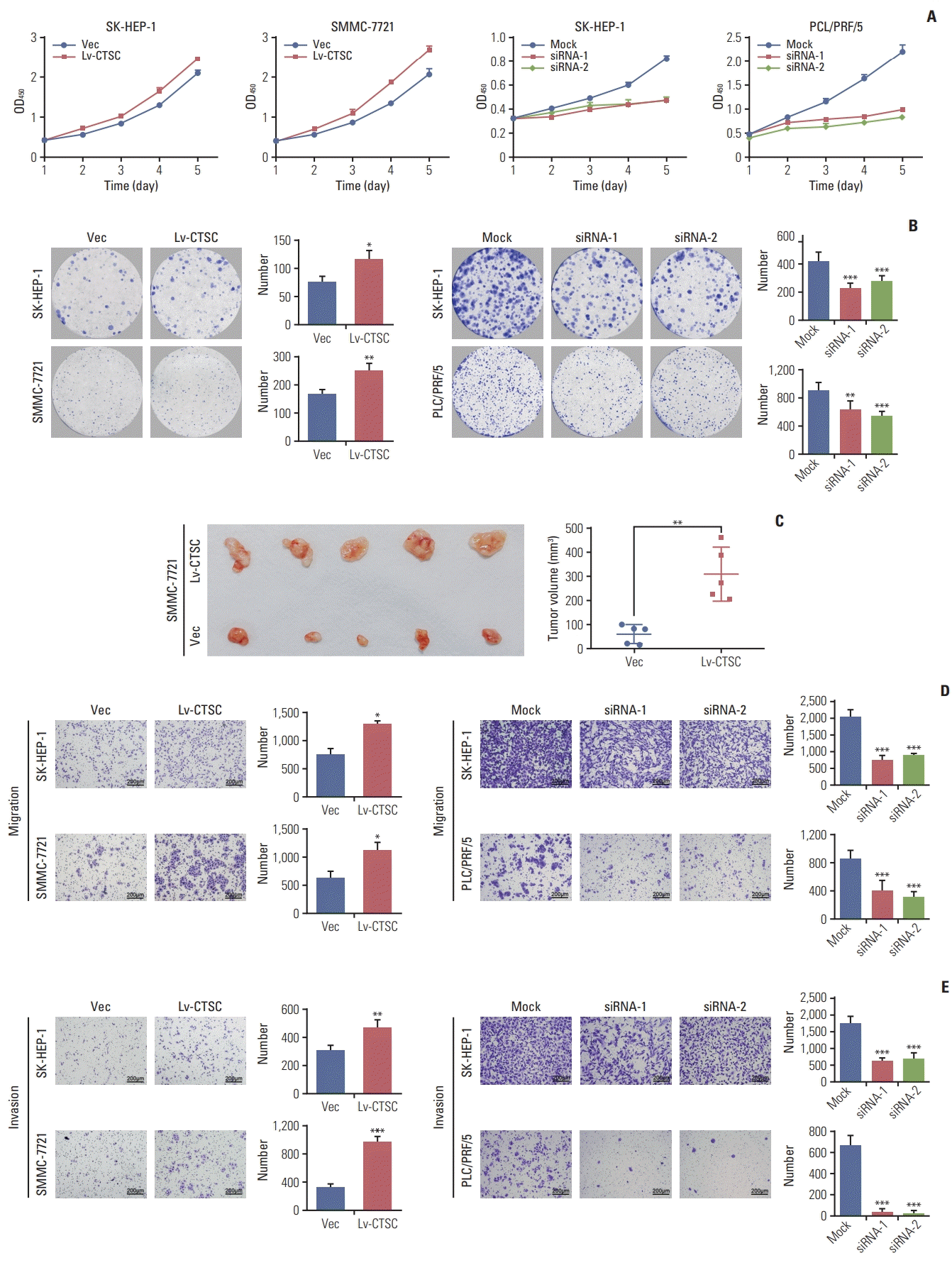 | Fig. 3.Cathepsin C (C TSC) promotes proliferation, migration, an d in vasion of hepatocellular carcinoma (HCC) cell lines. (A) The proliferation of SH-HEP-1 and SMMC-7721 cells infected with lentiviral LV-C TSC vector, or SK-HEP-1 and PLC/PRF/5 cells transfected with CTSC siRNA were assessed by Cell Counting Kit-8 assay. (B) The effects of CTSC on viability was assessed by colony formation assays. (C) Effects of overexpressing CTSC on tu morigenesis in vivo. (D) Trans-well chamber assays indicated the migratory ability of HCC cell lines. (E) Boyden chamber assays indicated thein vasiveness of HCC cell lines. *p < 0.05, **p < 0.01, ***p < 0.001. |
3. TNF-α/p38 MAPK pathway is involved in CTSC mediated HCC progression
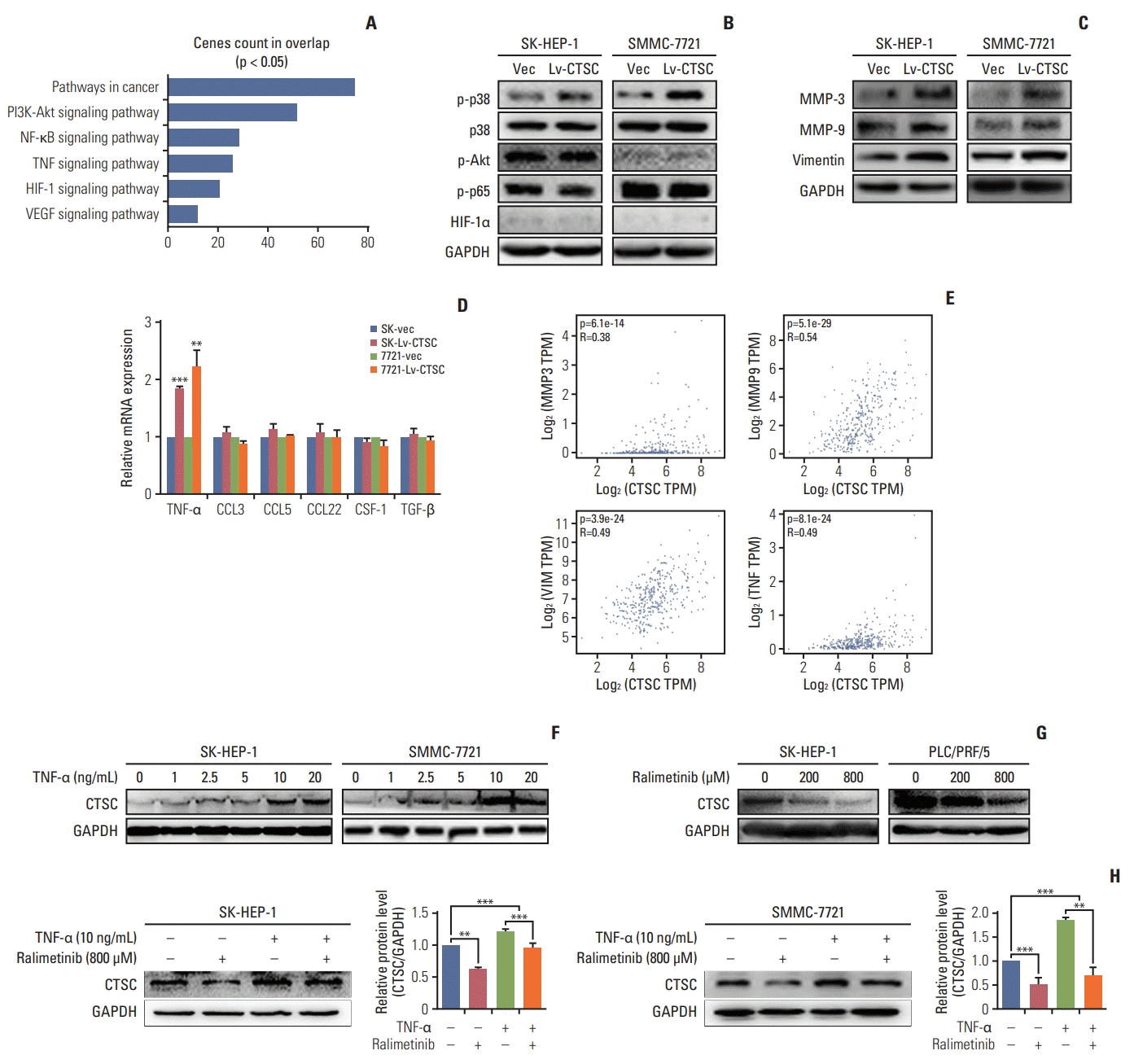 | Fig. 4.Tumor necrosis factor α (TNF-α)/mitogen-activated protein kinase (MAPK, p38) pathway is involved in cathepsin C (CTSC) mediated hepatocellular carcinoma progression. (A) The signaling pathway acquired by Gene Set Enrichment Analysis. (B) The activation of TNF-α/MAPK (p38) signaling pathway after overexpressing CTSC in SK-HEP-1 and SMMC-7721 cells, demonstrated by western blot assay. (C) The expression of matrix metalloproteinase (MMP) 3, MMP-9, vimentin after CTSC overexpression in SK-HEP-1 and SMMC-7721 cells detected by western blot assay. (D) The expression of TNF-α after overexpressing CTSC in SK-HEP-1 and SMMC-7721 cells detected by quantitative real-time polymerase chain reaction. (E) The correlation between CTSC and MMP-3, MMP-9, Vimentin, and TNF-α in clinical specimens analyzed by the GEPIA. (F) The western blot assay was used to investigate the effect of TNF-α on CTSC expression. (G, H) The western blot assay was used to investigate the effect of Ralimetinib on CTSC expression. PI3K, phosphoinositide 3-kinase; NF-κB, nuclear factor κB; TNF-α, tumor necrosis factor α; HIF-1, hypoxia-inducible factor 1; VEGF, vascular endothelial growth factor; GAPDH, glyceraldehyde 3-phosphate dehydrogenase. **p < 0.01, ***p < 0.001. |




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download