Abstract
Background
Carbapenem-resistant Enterobacteriaceae (CRE) with acquired metallo β-lactamase (MBL) resistance have been increasingly reported worldwide and associated with significant mortality and morbidity. Here, an outbreak of genetically related strains of Klebsiella pneumoniae producing the imipenemase (IMP)-1 MBL in a medical intensive care unit (MICU) in Korea is reported.
Methods
Since isolating carbapenem-resistant K. pneumoniae (CRKP) at the MICU of the hospital on August 10, 2011, surveillance cultures for CRE in 31 hospitalized patients were performed from August to September 2011. Carbapenem resistance was determined based on the disk diffusion method outlined in the Clinical and Laboratory Standards Institute guidelines. Polymerase chain reaction (PCR) was performed for genes coding for β-lactamase. Associations among isolates were assessed via pulsed-field gel electrophoresis (PFGE). In addition, a surveillance study of environmental cultures and health-care workers (HCWs) was conducted in the MICU during the same time frame.
Results
During the study period, non-duplicated CRKP specimens were discovered in four patients in the MICU, suggestive of an outbreak. On August 10, 2011, CRKP was isolated from the sputum of a 79-year-old male patient who was admitted to the MICU. A surveillance study to detect additional CRE carriers by rectal swab revealed an additional three CRKP isolates. PCR and sequencing of the four isolates identified the presence of the IMP-1 gene. In addition, PFGE showed that the four isolated strains were genetically related. CRE was not identified in specimens taken from the hands of HCWs or other environmental sources during surveillance following the outbreak. Transmission of the carbapenemase-producing Enterobacteriaceae strain was controlled by isolation of the patients and strict contact precautions.
Klebsiella pneumoniae is one of the major causes of nosocomial intraperitoneal, urinary tract, and primary bacteremia infections [1]. While carbapenem antibiotics are fairly stable against β-lactamases in the Enterobacteriaceae, the numbers of carbapenem resistant gram-negative strains are on the rise. The most common mechanism of carbapenem resistance in gram-negative bacteria is the production of β-lactamases [2,3] that incite carbapenem hydrolysis, most commonly through the production of metallo β-lactamases (MBLs) and class A and class D enzymes [4,5]. Imipenemase (IMP)-1 was the first identified MBL [6], and plasmid-mediated class B β-lactamases are classified into three major molecular groups: Verona integron-encoded metallo β-lactamase (VIM), IMP, and Sao Paulo MBL. IMP and VIM MBLs are increasingly reported in Asia, Europe, and America [7,8], and MBL has been sporadically reported in K. pneumoniae. IMP-1 β-lactamase produced by K. pneumoniae has been reported in Brazil [9], Singapore [10], and Japan [11], while IMP-8 production by K. pneumoniae has been reported in Taiwan [12].
In South Korea, carbapenem resistance in both Escherichia coli and K. pneumoniae remained below 1% until 2009, and there have been sporadic reports of carbapenemase-producing Enterobacteriaceae since then [13]. In Korea, sentinel surveillance of 6 multidrug resistant pathogens, including carbapenem-resistant Enterobacteriaceae (CRE), has been performed via revision of a precautionary measure of communicable diseases. In 2011, 26 cases of carbapenemase-producing Enterobacteriaceae (CPE) were identified: New Delhi metallo β-lactamase (NDM)-1 (8), IMP (7), and Klebsiella pneumoniae carbapenemase (KPC)-2 (5) [14]. 174 cases of CPE were reported in 2014. Among them, the most common Enterobacteriaceae was K. pneumoniae (104, 59.8%), and the carbapenemase was OXA-232 (48, 27.6%). Two cases of IMP-1 were reported (1.1%) [15]. There was a report of an outbreak of IMP-1 β-lactamaseproducing A. baumannii involving two cases of infection in Busan Korea in 2004 [16].
The present study aimed to report the epidemiological, microbiological, and clinical properties of an IMP-1-producing K. pneumoniae outbreak in the medical intensive care unit (MICU) of one healthcare institution.
The hospital of this study is a 947-bed tertiary hospital and the MICU contains 9 beds, including 2 isolated wards. After the first report of isolation of carbapenem-resistant K. pneumoniae (CRKP) at the MICU of the hospital on August 10, 2011, surveillance cultures to identify the spread of CRE were performed in 31 hospitalized patients from August to September 2011. Infection control interventions were implemented, and isolates were characterized by molecular epidemiology.
Information from CRKP patients was retrospectively collected through medical records, including patient age, gender, underlying disease, and clinical course. Specimen cultures were collected via a rectal swab. There were 31 patients in MICU who contacted the index patient. After notification of CRKP isolation, surveillance cultures were performed in 19 patients, while 12 patients were excluded because of status of discharge or death when notification of CRKP isolation occurred. At notification of the second CRE isolation, point surveillance was performed in patients staying in the MICU, and active surveillance by rectal swabs was performed in all patients newly admitted to the MICU. Patients who had a clinical or surveillance culture that was positive for CRE were contact isolated and received weekly cultures by rectal swab. In addition, surveillance cultures for both hands of all 26 MICU staff and the surrounding environment were performed. Environmental assessment was performed by swabbing various places within the MICU, including the fl oor, bed, monitor, and oxygen supply equipment. The acquired specimens were cultured in imipenem-containing MacConkey agar media. Polymerase chain reaction (PCR) and pulsed-field gel electrophoresis (PFGE) were conducted with the rectal swabs, environmental cultures, hand swabs, and carbapenemase genes to verify molecular genetic properties. In the study period, molecular characterization of CRE from clinical and surveillance specimens was carried out. Patients who had a clinical culture that was positive for CRE were screened for carriage of CRE. Rectal swab specimens were obtained for screening on the first day for the patients that had CRE detected via clinical culture.
After the initial CPE colonizations were recognized, an infection-control investigation was initiated and interventions were implemented. Infection–control interventions included temporary unit closure, use of cohort and contact isolation precautions for colonized patients, handwashing facilities, intensive cleaning of rooms and the unit of colonized patients (as a second step in the cleaning process), unit-targeted education of hospital staff, and re-enforcement of existing infection control policies. Patient- and room-specific procedures were continued until hospital discharge.
CRE was defined as Enterobacteriaceae that is nonsusceptible to one or more of the following carbapenems: doripenem, meropenem, ertapenem, or imipenem. All Enterobacteriaceae specimens were identified via the BD Phoenix automated system ID64 card (Becton Dickinson Diagnostic Systems), and carbapenem resistance was determined via the BD Phoenix automated system NMIC card (Becton Dickinson Diagnostic Systems, Sparks, MD, USA); this is based on the disk diffusion method recommended by the Clinical and Laboratory Standards Institute criteria based on their 2010 recommendations, which were in use in the laboratory during the time of this study [17]. The minimal inhibitory concentrations for imipenem and meropenem were assessed via Etest® (bioMérieux, Durham, NC, USA). Each specimen was assessed for carbapenemase production using the modified Hodge test (MHT) and the carbapenemase inhibition test was performed using the KPC + MBL confirm ID kit (Rosco Diagnostica, Copenhagen, Denmark), with meropenem 10 μg (MRP10), meropenem 10 μg + dipicolinic acid (metallo β-lactamase inhibitor; MR+DP), meropenem 10 μg + boronic acid (KPC and AmpC inhibitor; MR+BO), and meropenem 10 μg + cloxacillin (AmpC inhibitor; MR + CX).
For specimens that were positive for Enterobacteriaceae, PCR was performed to identify the NDM-1, KPC, IMP, and VIM genes. PCR was conducted using a Mastercycler 384 instrument (Eppendorf Scientific Inc., Hamburg, Germany) under the following conditions: denaturation for 5 min at 94°C, 30 cycles of 30 sec at 94°C, 30 sec at 56°C, 30 sec at 72°C, and a final extension for 7 min at 72°C. The nucleic acid sequence was analyzed using the Basic Local Alignment Search Tool provided by the National Center for Biotechnology (Table 1).
Associations among strain isolates were assessed via XbaI PFGE. The bacterial DNA was cut with XbaI (New England Biolabs, Beverly, MA, USA) and isolated in 1.2% agarose gel (in 0.5× TBE buffer) using the CHEF Mapper apparatus (Bio-Rad Laboratories, Hercules, CA, USA).
Statistical analysis was performed using IBM SPSS Statistics for Windows, version 23.0 (IBM Corp., Armonk, NY, USA). Correlations between CRE carriers and non-CRE carriers were evaluated using Fisher’s exact test, and continuous variables were evaluated using Student’s t-test. P-values less than 0.05 were considered statistically significant.
During the study period, non-duplicated CRKP specimens were discovered in four patients in the MICU, suggestive of an outbreak. Figure 1 shows the CPE outbreak over time at the MICU. It was difficult to determine whether patient 1 or patient 2 was the index case of this outbreak.
A total of 19 patients were involved in the study and received clinical and surveillance cultures. Surveillance cultures of rectal swabs (19) and clinical cultures of blood (19), sputum (9), urine (5), and wound (3) specimens were analyzed. Positive cultures for CRE included one sputum, one wound specimen from a tracheostomy site, and four rectal swabs; all were CPKP.
Patient 1, the first reported patient with CPE, was a 79-year-old male patient transferred to the hospital and admitted to the MICU. He had a history of hypertension and gout, and received mechanical ventilation upon diagnosis with pneumonia. He received piperacillin/tazobactam and levofl oxacin for 10 days from day 1 of hospitalization, and carbapenemase-producing K. pneumoniae that did not produce extended spectrum β-lactamase was identified on day 11 from sputum acquired from the endotracheal tube. No additional treatment was administered for this strain of bacteria as the pneumonia showed improvement. On the day the identification of carbapenem-resistant K. pneumoniae was notified (day 8 after the initial test), surveillance rectal swab cultures for CRE were conducted in three other patients whose hospital stays overlapped with patient 1. Six days following the test, one out of three patients (patient 2) tested positive for CPE. A CPE outbreak was declared. Further admission into the MICU was prohibited and infection management for CPE was began. Strict contact precautions were enforced, including contact isolation precautions for colonized patients, hand washing facilities, and education of hospital staff. Intensive cleaning of the rooms and unit of colonized patients were performed (as a second step in the cleaning process), and re-enforcement of existing infection control policies were performed.
Patient 2 was a 79-year-old female patient with a cerebral infarction and femoral neck fracture who was in the MICU for 42 days. The first 13 days overlapped with patient 1. Similar to patient 1, she was transferred to the hospital and received piperacillin/sulbactam and ciprofloxacin treatment for acute pyelonephritis for 17 days prior to the rectal swab. Patient 2 was transferred to a long-term care hospital upon improvement.
On the day the isolation of CPE from the rectal swab of patient 2 was notified, surveillance rectal swab cultures for 15 patients (of these, six were admitted to the MICU at the time) who were not tested were performed. On day 10 of the surveillance swabs, CRE was identified in one out of 15 patients (patient 3). Patient 3 was a 72-year-old female patient with primary peritoneal cancer who received cefoperazone/sulbactam treatment after being diagnosed with rhabdomyolysis. She was transferred to a different hospital after 9 days in the MICU, and her entire stay at the MICU overlapped with patient 2. There were three patients in the MICU on the day CPE was identified; after isolating patient 2, who had tested positive for CPE, new admissions into the MICU were permitted. From this point, active surveillance cultures (ASCs) for CRE on all new patients in the MICU were performed via rectal swabs. On the 34th day of ASCs, CPE was identified in the tracheostomy wound site of patient 4, a 64-year-old female patient who had been receiving care for 39 days in the MICU. In the MICU, patient 4 overlapped for 26 days with patient 2 and 8 days with patient 3. Her rectal swab surveillance culture on day 8 of her hospitalization was negative for CRE, but CPE was identified from the tracheostomy wound site conducted 46 days later. She was given ceftriaxone and vancomycin for pneumonia, and expired on the 111th day of admission from a cause unrelated to the isolated CPE. Vancomycin-resistant enterococcus was also isolated from her rectal specimen. All four patients who tested positive for CPE were supported by endotracheal intubation, mechanical ventilation, central vein cannulation, and an indwelling urinary catheter (Table 2). The beds of patients with overlapping MICU stays were not adjacent and were separated by another bed.
Antibiotic susceptibility tests for the strain isolates from these four patients revealed that all of the CPE strains were resistant to different types of antibiotics, with the exception of amikacin and gentamicin (Table 3). MBL production was confirmed in the isolated strains, and PCR and nucleic acid sequence analysis confirmed the presence of the blaIMP-1 gene. All of the four isolated strains were MHT positive (Table 4). In addition, PFGE showed that the four isolated strains were genetically related (Figure 2). CPE was not identified in the specimens taken from the hands of health-care workers or other environmental sources during environmental surveillance following the outbreak. The outbreak was terminated after all colonized patients of CPE were transferred to a separate isolation unit and there was no additional CRE identification for two weeks after the last reported case. Three negative perianal cultures obtained at weekly intervals were suggested as a criterion for discontinuation of contact precautions. Surveillance rectal swab cultures for all newly admitted patients at the MICU were performed, and there were no additional CRE identifications. Active surveillance culture for CRE by rectal swab has continued for all patients admitted to the MICU to date.
In the analysis of risk factors for CRE isolation, there were no significant differences compared to non-CRE carriers (Table 5). Transfer from another hospital, underlying diseases (diabetes, chronic liver disease, malignancy, and heart failure), use of medical devices, and antibiotics therapies were analyzed. Final outcomes and hospital days were also compared between CRE carriers and non-CRE carriers.
All four patients who tested positive for CPE received piperacillin or cephalosporin, not carbapenem, before isolation of MBL-producing CRE. There have been reports that in addition to β-lactams, several types of antibiotics, including fluoroquinolone and vancomycin, have been associated with CRE carrier status and infection [ 18-20]. Therefore, antibiotic management focused on the reduction of overall use of antibiotics, as opposed to a focus on specific antibiotics, may be the most effective method to reduce the incidence of CPE. In addition, given that the four patients were hospitalized for an average of 22 days at the MICU and total hospital stay would have been longer because they were transferred to the hospital, the use of a central vein cannula, indwelling urinary catheters, and mechanical ventilation may also be related to CPE infection [19]. Other studies have also reported that CRE was confirmed from medical equipmentrelated infections, particularly catheter-related urinary tract infections [21]. As use of medical equipment is an easy mediator for pathogen colonization and infections, limiting the use of invasive equipment may be another important method of preventing CPE infection [21]. Patel et al. [19] reported that CRKP infection and colonization are more highly associated with longer hospital stays for patients who have received solid organ or stem cell transplantation and are on mechanical ventilation, as compared to carbapenem-sensitive K. pneumoniae infection and colonization. Malnutrition and treatment in the ICU are other known risk factors for CRKP [20].
In this study, there was no significant risk factor for CRE carriage. Limitations include the small number of subjects and the fact that all were admitted to the MICU. Further well-designed studies of CRE carriage/infection should be done to establish best practices for prevention of CRE transmission.
In the present study, CPE was not isolated from environmental cultures, including the hands of medical staff. This may be due to temporary or short-term colonization [22]; however, the exact mechanism of transmission could not be confirmed. Several studies have suggested that isolated strains frequently contaminate the surrounding environment during an outbreak [23,24], and some mutant strains may survive for months, even on dry surfaces [25,26]. Medical staff are well known vectors of resistant bacterial strains, where bacteria are transmitted through the hands or clothes [27]. The fact that the beds of the CPE-positive patients were not adjacent supports the possibility of transmission by medical staff.
The clinical data and PFGE analysis results showed that the MBL-producing K. pneumoniae isolates from four patients were related, suggesting this outbreak was a result of a nosocomial infection. Furthermore, the four patients had overlapped stays at the MICU, during which the transmission may have occurred. Additional MBL-producing K. pneumoniae was not identified, owing to the implementation of measures such as patient isolation, prohibition of new admissions, and medical staff training regarding infection control (e.g., hand hygiene). The four patients in this study did not show actual infections; rather, they had asymptomatic colonization of CPE, which may have been cross transmitted through the hands of the medical staff or general environmental factors. CPE was initially detected at the hospital but could also have been transmitted to the local community, as Enterobacteriaceae is a common cause of nosocomial and local community infections [28].
Stringent infection management via active surveillance and patient isolation is required to prevent and prepare for the transmission and spread of resistance bacteria throughout hospitals and communities. Furthermore, laboratories should strive to quickly diagnose the cause and monitor through continuous surveillance. Immediately verifying the occurrence of CPE in the clinical laboratory is the first step towards prevention, and it is important for medical institutions to understand how common CPE is in their institutions. According to a report by the Centers for Disease Control, failure of early recognition of the isolation of CPE within an institution is a risk factor for widespread distribution of the bacteria. Patients who test positive for CPE must be treated with contact precaution, and some experts even argue that these patients should be cohorted [29]. In addition, if CPE infection is confirmed through a culture or point prevalence survey, the corresponding institution must consider performing surveillance cultures for patients that are epidemiologically related to the isolated patients [30]. The purpose of such cultures is to verify additional unrecognized patients with CPE colonization who can potentially transmit the bacteria.
In conclusion, the present surveillance culture study of an IMP-1-producing K. pneumoniae outbreak in an ICU suggests that it is important to monitor the progression of colonization or infection through rectal swab tests and environmental assessments. As CPE can pose threats not only to a single institution but also the entire local community, the role of public healthcare is especially important for the prevention of CPE, and occurrences of CPE, including carbapenemases, should be the subject of continuous monitoring.
References
1. Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998; 11:589–603.
2. Nikaido H. Prevention of drug access to bacterial targets: permeability barriers and active effl ux. Science. 1994; 264:382–8.
3. Piddock LJ. Clinically relevant chromosomally encoded multidrug resistance effl ux pumps in bacteria. Clin Microbiol Rev. 2006; 19:382–402.
6. Watanabe M, Iyobe S, Inoue M, Mitsuhashi S. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1991; 35:147–51.
7. Nordmann P, Poirel L. Emerging carbapenemases in Gram-negative aerobes. Clin Microbiol Infect. 2002; 8:321–31.

8. Walsh TR, Toleman MA, Poirel L, Nordmann P. Metallo-beta-lactamases: the quiet before the storm? Clin Microbiol Rev. 2005; 18:306–25.
9. Lincopan N, McCulloch JA, Reinert C, Cassettari VC, Gales AC, Mamizuka EM. First isolation of metallo-beta-lactamase-producing multiresistant Klebsiella pneumoniae from a patient in Brazil. J Clin Microbiol. 2005; 43:516–9.
10. Koh TH, Babini GS, Woodford N, Sng LH, Hall LM, Livermore DM. Carbapenem-hydrolysing IMP-1 beta-lactamase in Klebsiella pneumoniae from Singapore. Lancet. 1999; 353:2162.
11. Shibata N, Doi Y, Yamane K, Yagi T, Kurokawa H, Shibayama K, et al. PCR typing of genetic determinants for metallo-beta-lactamases and integrases carried by gram-negative bacteria isolated in Japan, with focus on the class 3 integron. J Clin Microbiol. 2003; 41:5407–13.
12. Yan JJ, Ko WC, Tsai SH, Wu HM, Wu JJ. Outbreak of infection with multidrug-resistant Klebsiella pneumoniae carrying blaIMP-8 in a university medical center in Taiwan. J Clin Microbiol. 2001; 39:4433–9.
13. Centers for Disease Control and Prevention. Monitoring of antimicrobial resistance on general hospitals in Korea [Internet]. Cheongju: Centers for Disease Control and Prevention;c2010. [cited 2010 Aug 27]. Available from: http://cdc.go.kr/CDC/info/CdcKrInfo0301.jsp?menuIds=HOME001-MNU1154-MNU0005-MNU0037-MNU1380&cid=12505.
14. Yoo JS. Emergence and charcteristics of Carbapenemase-Producing Enterobacteriaceae(CPE) in Korea, 2011 [Internet]. Cheongju: Centers for Disease Control and Prevention;c2012. [cited 2012 May 4]. Available from: http://cdc.go.kr/CDC/cms/cmsFile-Download.jsp?fid=31&cid=20994&fieldName=attach1&index=1.
15. Park JW, Lee EJ, Lee D. Status of Carbapenemase producing Enterobacteriaceae in Korea, 2014 [Internet]. Cheongju: Centers for Disease Control and Prevention;c2015. [cited 2015 Dec 31]. Available from: http://cdc.go.kr/CDC/info/CdcKrInfo0301.jsp?menuIds=HOME001-MNU1154-MNU0005-MNU0037-MNU1380&cid=66669.
16. Jeong SH, Bae IK, Park KO, An YJ, Sohn SG, Jang SJ, et al. Outbreaks of imipenem-resistant Acinetobacter baumannii producing carbapenemases in Korea. J Microbiol. 2006; 44:423–31.
17. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: Twentieth Informational Supplement M100-S20. Wayne: Clinical and Laboratory Standards Institute;2010.
18. Hussein K, Sprecher H, Mashiach T, Oren I, Kassis I, Finkelstein R. Carbapenem resistance among Klebsiella pneumoniae isolates risk factors, molecular characteristics, and susceptibility patterns. Infect Control Hosp Epidemiol. 2009; 30:666–71.
19. Patel G, Huprikar S, Factor SH, Jenkins SG, Calfee DP. Outcomes of carbapenem-resistant Klebsiella pneumoniae infection and the impact of antimicrobial and adjunctive therapies. Infect Control Hosp Epidemiol. 2008; 29:1099–106.
20. Schwaber MJ, Klarfeld-Lidji S, Navon-Venezia S, Schwartz D, Leavitt A, Carmeli Y. Predictors of carbapenem-resistant Klebsiella pneumoniae acquisition among hospitalized adults and effect of acquisition on mortality. Antimicrob Agents Chemother. 2008; 52:1028–33.
21. Gould CV, Umscheid CA, Agarwal RK, Kuntz G, Pegues DA; Healthcare Infection Control Practices Advisory Committee. Guideline for prevention of catheter-associated urinary tract infections 2009. Infect Control Hosp Epidemiol. 2010; 31:319–26.

22. Chu YW, Leung CM, Houang ET, Ng KC, Leung CB, Leung HY, et al. Skin carriage of acinetobacters in Hong Kong. J Clin Microbiol. 1999; 37:2962–7.

23. Tankovic J, Legrand P, De Gatines G, Chemineau V, Brun-Buisson C, Duval J. Characterization of a hospital outbreak of imipenem-resistant Acinetobacter baumannii by phenotypic and genotypic typing methods. J Clin Microbiol. 1994; 32:2677–81.
24. Go ES, Urban C, Burns J, Kreiswirth B, Eisner W, Mariano N, et al. Clinical and molecular epidemiology of acinetobacter infections sensitive only to polymyxin B and sulbactam. Lancet. 1994; 344:1329–32.

25. Catalano M, Quelle LS, Jeric PE, Di Martino A, Maimone SM. Survival of Acinetobacter baumannii on bed rails during an outbreak and during sporadic cases. J Hosp Infect. 1999; 42:27–35.
26. Wendt C, Dietze B, Dietz E, Rüden H. Survival of Acinetobacter baumannii on dry surfaces. J Clin Microbiol. 1997; 35:1394–7.
27. Wilson AP, Smyth D, Moore G, Singleton J, Jackson R, Gant V, et al. The impact of enhanced cleaning within the intensive care unit on contamination of the near-patient environment with hospital pathogens: a randomized crossover study in critical care units in two hospitals. Crit Care Med. 2011; 39:651–8.

28. Bratu S, Landman D, Haag R, Recco R, Eramo A, Alam M, et al. Rapid spread of carbapenem-resistant Klebsiella pneumoniae in New York City: a new threat to our antibiotic armamentarium. Arch Intern Med. 2005; 165:1430–5.
29. Bilavsky E, Schwaber MJ, Carmeli Y. How to stem the tide of carbapenemase-producing enterobacteriaceae?: proactive versus reactive strategies. Curr Opin Infect Dis. 2010; 23:327–31.

30. Centers for Disease Control and Prevention (CDC). Guidance for control of infections with carbapenem-resistant or carbapenemase-producing Enterobacteriaceae in acute care facilities. MMWR Morb Mortal Wkly Rep. 2009; 58:256–60.
Figure 1.
Timeline for the described CRE outbreak in the MICU, 2011. CRE: carbapenem-resistant Enterobacteriaceae; MICU: medical intensive care unit; VRE: vancomycin-resistant enterococcus; PFGE: pulsed-field gel electrophoresis.
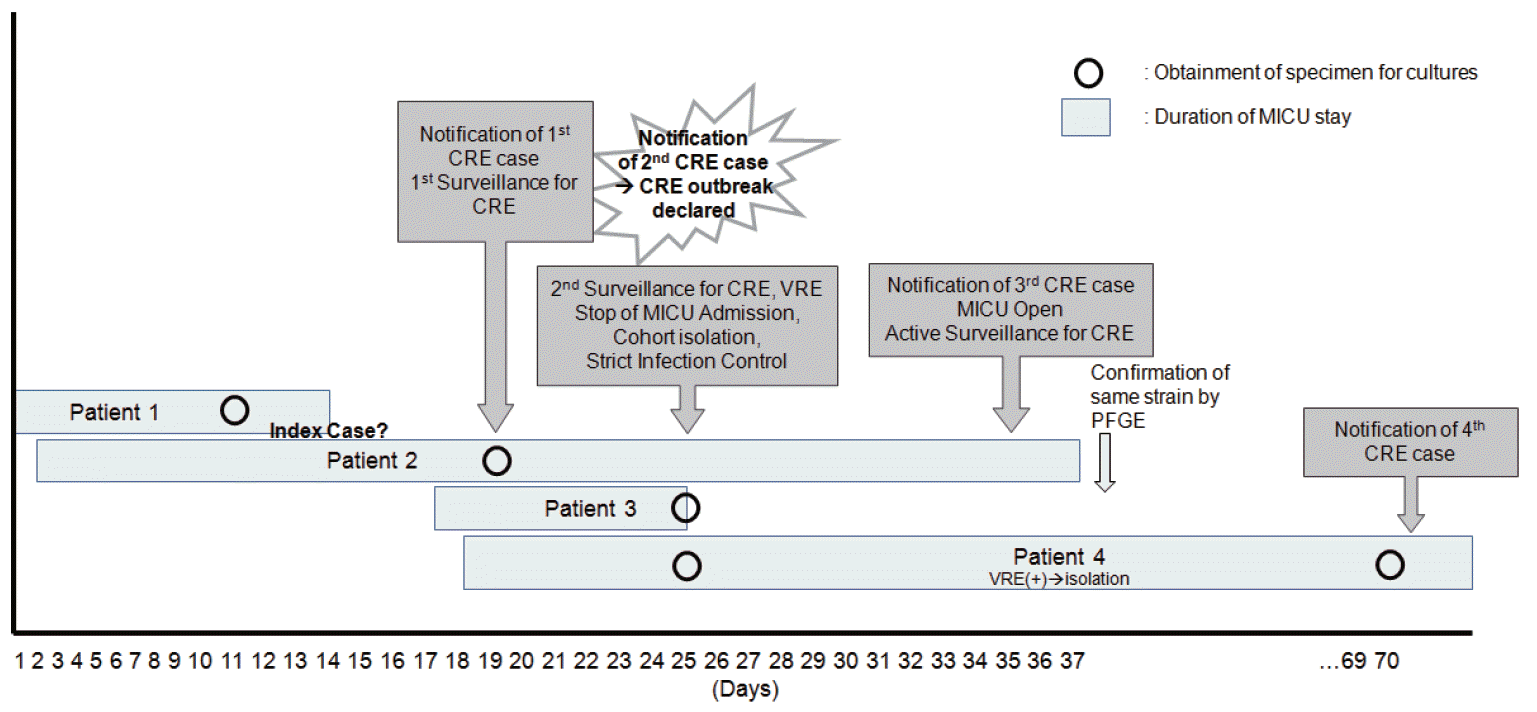
Figure 2.
Dendrogram of PFGE macro restriction patterns generated with the RFLPrint computer software. The scale indicates percent similarity. PFGE profiles of K. pneumoniae strains isolated from 4 patients in the MICU. PFGE: pulsed-field gel electrophoresis; MICU: medical intensive care unit.
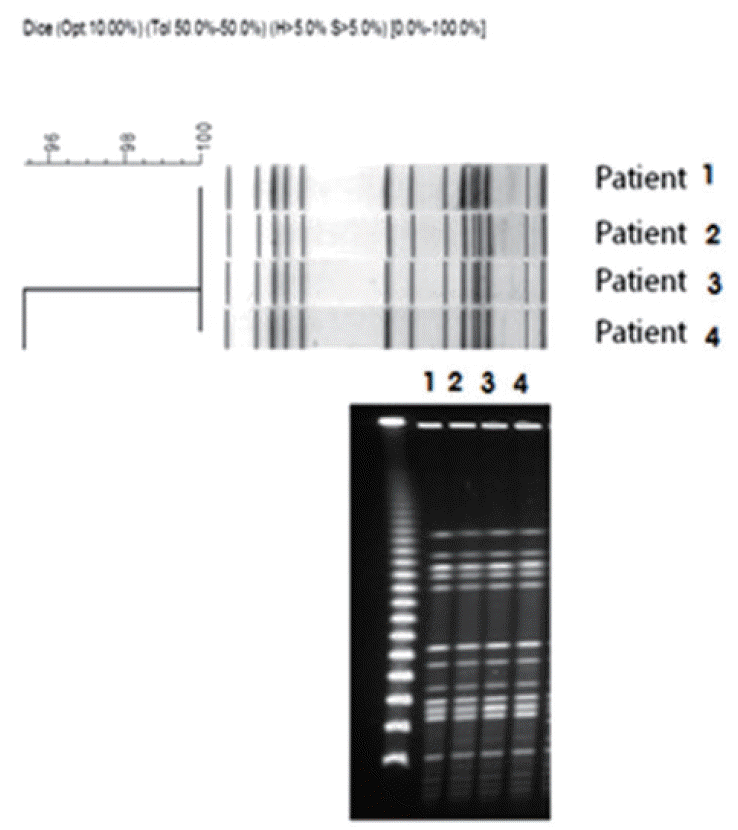
Table 1.
Nucleotide sequences of oligonucleotides used for PCR amplification and sequencing of VIM, IMP, NDM, and KPC genes
Table 2.
Clinical characteristics of 4 patients with carbapenem resistant K. pneumoniae isolates
Table 3.
Antimicrobial susceptibility profile of 4 carbapenem resistant K. pneumoniae isolates
Table 4.
Laboratory characteristics of 4 carbapenem resistant K. pneumoniae isolates
Table 5.
Risk factors associated with CRE carriers




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download