Abstract
Purpose
The purpose of this study was to investigate the prognostic significance of liver kinase b1 (LKB1) loss in patients with operable colon cancer (CC).
Materials and Methods
Two hundred sixty-two specimens from consecutive patients with stage III or high-risk stage II CC, who underwent surgical resection with curative intent and received adjuvant chemotherapy with fluoropyrimidine and oxaliplatin, were analyzed for LKB1 protein expression loss, by immunohistochemistry as well as for KRAS exon 2 and BRAFV600E mutations by Sanger sequencing and TS, ERCC1, MYC, and NEDD9 mRNA expression by real-time quantitative reverse transcription polymerase chain reaction.
Results
LKB1 expression loss was observed in 117 patients (44.7%) patients and correlated with right-sided located primaries (p=0.032), and pericolic lymph nodes involvement (p=0.003), BRAFV600E mutations (p=0.024), and TS mRNA expression (p=0.041). Patients with LKB1 expression loss experienced significantly lower disease-free survival (DFS) (hazard ratio [HR], 1.287; 95% confidence interval [CI], 1.093 to 1.654; p=0.021) and overall survival (OS) (HR, 1.541; 95% CI, 1.197 to 1.932; p=0.002), compared to patients with LKB1 expressing expressing tumors. Multivariate analysis revealed LKB1 expression loss as independent prognostic factor for both decreased DFS (HR, 1.217; 95% CI, 1.074 to 1.812; p=0.034) and decreased OS (HR, 1.467; 95% CI, 1.226 to 2.122; p=0.019).
Colorectal cancer (CRC) causes high morbidity and mortality rates; however, both declined the last four decades in Western countries [1]. Cancer prevention could be attributed through screening at an early stage and more effective treatment modalities. Moreover, it was shown that patients with high-risk stage II and stage III colon cancer (CC) who received the current standard treatment according to the National Comprehensive Cancer Network guidelines had a benefit on their survival rates [2].
For stage III disease, combination chemotherapy with a backbone of fluoropyrimidine and oxaliplatin (FOLFOX, CAPOX) is the current standard of care, since it leads to prolongation of both disease-free survival (DFS) and overall survival (OS) [3]. The results for the addition of oxaliplatin in OS and DFS for patients with stage II CC with high-risk features (such as T4 tumors, obstruction or perforation, and vessel invasion) showed a marginal but significant [3]. Despite that, it has long been recognized patients’ individual risk of recurrence varies widely even in patients with the same stage in CC. Since today, microsatellite instability (MSI) status is the only biomarker used in daily clinical practice [4].
Several studies have reported that germline inactivating mutations in liver kinase b1 (LKB1) to be the primary cause syndromes, such as the Peutz-Jeghers syndrome, which confer an increased risk of cancer development [5,6]. The LKB1 also known as serine threonine kinase 11 (STK11), was initially discovered as a tumor suppressor gene and it has been implicated on initiation and progression of neoplastic diseases [5]. It controls a wide range of diverse cellular processes through phosphorylation of the adenosine monophosphate-activated protein kinase (AMPK) protein when cellular energy levels are depleted promoting ATP [6]. Furthermore, loss of LKB1 has been shown to influence cell polarity, epi-thelial-to-mesenchymal transition, apoptosis, angiogenesis, and cell cycle inhibition [7]. Also, negatively regulates the mammalian target of rapamycin signaling [5] and mediates p53 activation [8]. LKB1 has been most intensively studied using lung cancer mouse models [9]. Finally, the results of a first comprehensive meta-analysis suggested that decreased LKB1 expression significantly contributed to shorter OS in solid tumor patients [10]. However, additional studies rela-ted to specific tumor types and perspectives are required to verify the clinical utility of decreased LKB1 levels in solid tumors.
Based on the above-mentioned data we conducted a retrospective biomarkers-based studies, in order to elucidate the clinicopathological features and prognostic significance of LKB1 loss of expression in high-risk stage II and stage III CC, treated with oxaliplatin and fluoropyrimidine combination adjuvant chemotherapy.
Two hundred and two formalin-fixed, paraffin-embedded samples from consecutive patients with stage III or high-risk stage II CC treated with FOLFOX or CAPOX has been studied.
The most enriched in cancer cells areas were selected by a pathologist (M.T.) and afterwards, serial sections of 5 μm were stained by nuclear Fast Red (Sigma-Aldrich, St. Louis, MO). Micro-dissection, using a piezoelectric micro-dissector (Eppendorf, Hamburg, Germany) was performed in cases with lower than 80% of neoplastic cells in the examined section. Extraction of nucleic acids (DNA and RNA) was performed, according to the manufacturer’s protocol as previously described [11].
KRAS exon 2 and BRAFV600E mutation mutations analysis was carried out by Sanger sequencing after polymerase chain reaction (PCR) amplification and by reverse transcription PCR using allelic discrimination method, respectively, as previously described as previously reported [11,12]. Analysis was performed using the SDS 2.3 software [13].
The MSI analysis was carried out with the use of Promega MSI Analysis System (Promega, Madison, WI) according to the manufacturer’s instructions. Amplicons for MSI detection was performed by capillary electrophoresis on an ABI 3130xl Genetic Analyzer following PCR amplification and analyzed using GeneMapper Software, ver. 3.7 (Applied Biosystems/Life Technologies, Grand Island, NY) [14].
Synthesis of cDNA was done as described previously [15]. Primers and probes were designed using the Primer Express 2.0 Software (Applied Biosystems) according to the Ref Seq NM_002467.4 for MYC-ERCC1-NEDD9-TS. The sets of primers probes are provided in S1 Table, while those for the housekeeping genes, β-actin and PGK have been previously published [13]. The quantification of mRNA expression was carried out using the 2−(ΔCT sample−ΔCT calibrator) method, as previously described [13]. Only triplicates with a standard deviation less than 0.25 were accepted.
We performed immunostain using Thermo Scientific UltraVision Quanto Detection System HRP and polyclonal antibody for LKB1 (1:100 dilution, Thermo Scientific, Waltham, MA). Adult seminiferous tubules of the testis were used as a positive control as it shows the highest levels of LKB1 expression [16]. Negative control was obtained by omitting the primary antibody. Immunohistochemistry (IHC) staining intensity was measured using a scaling system of 0 (no expression), 1, 2, and 3 (highest expression) blinded by a pathologist. A weighted index (WI) was applied in both the nucleus and the cytoplasm using the equation WI=% tumor stain X intensity score as used in another study [16]. Cytoplasmic, nuclear expression or both were considered a positive stain. The staining of LKB1 was interpreted without knowledge of its genetic status.
DFS was estimated as the interval between the date of colectomy to the first documented disease progression, second primary CC or death. OS was calculated from the date of surgery to date of death. The Kaplan-Meier survival curves were used for survival analysis and correlations with the studied parameters, while Cox proportional hazards model was applied to weigh the validity of considered factors on study defined events. The statistically significant factors were then incorporated in a multivariate Cox proportional hazards regression model in order to estimate their unbiased significance on progression or survival. The only p-value of < 0.05 was considered as significant.
The median values of mRNA expression were used as cut-off points, with samples above or equal to the median characterized as high expressing, while those with value below the median as low expression. The laboratory research was completed blinded to the clinical parameters. Correlations between the studied biomarker with baseline characteristics were calculating by Fisher exact test for categorical variables or logistic regression for continuous ones.
The study has been approved by the Ethics and Scientific Committees of the University General Hospital of Heraklion (number of approval: 2058) and written informed consent for the use of their tissue for translational research was obtained from all patients. All authors, declare no competing interest regarding this study.
The main demographic and clinical characteristics of the study population are summarized in Table 1. Briefly, patients with CC were predominately males (58%), with a median age of 67 years and the majority of them with a good performance status of 0-1. In addition, 60% of the patients were diagnosed with stage III colon cancer, 65% had primary tumor located in the left colon and 61% low-grade tumors (Table 1). Almost two-thirds of the patients (65%) received adjuvant treatment with CAPOX and the rest one third with FOLFOX. At the time of analysis and after a median follow-up of 120.7 months (min-max, 11.3 to 161.1 months); 71 (27%) disease relapses and 48 (18%) deaths have been recorded.
The results of the laboratory analysis are presented in Table 2. Analysis for LKB1 protein expression was successfully performed in all 254 specimens (96.9%), whereas KRAS exon 2 and BRAF exon 15 mutation analysis was performed in 251 (95.8%) and 246 (93.9%) specimens,respectively (Table 2, S2-S5 Figs.). Mismatch repair (MMR) system status was successfully analyzed in 235 (89.7%) specimens, while mRNA expression of ERCC1, MYC, NEDD9, and TS was successfully done in 247 (94.3%), 250 (95.4%), 249 (95%), and 226 (86.2%) specimens, respectively (S6 Fig.).
Loss of LKB1 protein expression was observed in 117 (44.7%) tumors and correlated significantly with pericolic lymph node involvement (p=0.003) and primary tumors located in the right colon (p=0.032), whereas no significant correlation of LKB1 protein expression with age, sex, and grade were found (Table 3). In addition, LKB1 protein expression loss was significantly correlated with BRAFV600E mutation (p=0.024), and TS mRNA (p=0.041). In contrast, LKB1 protein expression loss was not significantly associated with KRAS exon 2 mutations, MMR status or ERCC1, MYC, NEDD9 mRNA expression (all p > 0.05).
The correlations of analyzed markers and clinic-pathological features with DFS and OS are presented in Tables 4 and 5, respectively. Patients with tumors with LKB1 expression loss showed significantly lower DFS compared with those with LKB1 positive tumors (hazard ratio [HR], 1.287; 95% confidence interval [CI], 1.093 to 1.654; p=0.021) (Table 4, Fig. 1). In addition, patients with BRAFV600E mutations in their primary tumors presented higher probability for relapse compared to patients with BRAFV600E wild type tumors (HR, 1.976; 95% CI, 1.793 to 2.495; p=0.001) (Table 4). Likewise, patients with KRAS exon 2 mutations in their primary tumors presented higher probability for relapse compared to patients with KRAS exon 2 wild type tumors, but the difference was marginally significant (HR, 1.757; 95% CI, 1.000 to 3.090; p=0.05). Furthermore, patients with proficient MMR (pMMR) tumor had a significantly higher risk of relapse in comparison with those with deficient MMR (dMMR) tumors (HR, 1.726; 95% CI, 1.289 to 3.514; p=0.025). Finally, stage III disease at diagnosis is correlated with increased risk of progression compared with stage II (HR, 1.803; 95% CI, 1.605 to 2.055; p=0.023). All other comparisons between the mRNA expression of the ERCC1, MYC, NEDD9, and TS or several clinicopathological features, such as age, sex, tumor location and grade, did not reveal any significant correlations with DFS (all long-rank p > 0.05).
Regarding OS, patients with tumors with LKB1 expression loss showed significantly lower OS compared with those with LKB1 positive tumors (HR, 1.541; 95% CI, 1.197 to 1.932; p=0.002) (Table 4, Fig. 2). Similarly, patients with BRAFV600E mutations in their primary tumors presented higher probability for death compared to patients with BRAFV600E mutations wild type tumors (HR, 1.624; 95% CI, 1.143 to 2.309; p=0.007) (Table 4). In addition, patients with pMMR tumor had a significantly higher risk of death in comparison with those with dMMR tumors (HR, 1.375; 95% CI, 1.043 to 2.711; p=0.036). Also, stage III disease at diagnosis is correlated with increased risk of progression compared with stage II (HR, 1.636; 95% CI, 1.487 to 2.011; p=0.03). All other comparison did not reveal any significant correlations between the mRNA expression of the ERCC1, MYC, NEDD9, and TS or the detection of KRAS exon 2 mutation as well as clinicpathological features, such as age, sex, tumor location and grade, with DFS (all long-rank p > 0.05).
Multivariate analysis for DFS, revealed that LKB1 expression loss (HR, 1.217; 95% CI, 1.074 to 1.812; p=0.034) (Table 5), BRAFV600E mutations (HR, 1.696; 95% CI, 1.365 to 2.294; p=0.011), pMMR status (HR, 1.775; 95% CI, 1.343 to 3.011; p=0.007) and stage III disease (HR, 1.784; 95% CI, 1.335 to 2.762; p=0.006) as independent factors for increased risk of relapse. Similarly, LKB1 expression loss (HR, 1.467; 95% CI, 1.226 to 2.122; p=0.019), BRAFV600E mutations (HR, 1.961; 95% CI, 1.656 to 2.949; p=0.001), pMMR status (HR, 1.575; 95% CI, 1.243 to 3.001; p=0.018), and stage III disease (HR, 1.843; 95% CI, 1.356 to 2.623; p=0.009) was statistically associated with risk of death.
The data presented in the current study, demonstrated for the first time in literature, the impact of testing LKB1 protein expression in stage II/III CC patients that underwent resection and subsequently received CAPOX or FOLFOX adjuvant chemotherapy. Additionally, this retrospective study aims to identify correlations of LKB1 expression loss with known clinic-pathological features and common mutations (KRAS exon 2 and BRAFV600E) mutation in operable CC. Moreover, we analyzed the predictive significance of these biomarkers in conjunction with ERCC1, MYC, NEDD9, and TS mRNA expression. Based on the literature, this is the first study that correlates a combination of all these parameters, hence; the results of our analysis could potentially serve as an advantageous guide for every day clinical practice.
AMPK is greatly regulated by LKB1 activity and is vital for cell metabolism through the maintenance of energy homeostasis. LKB1 growth-suppressing effect is operated through activation of twelve AMPK-related kinases. This AMPK-related kinases activation by LKB1 is crucial for the regulation of (1) cell metabolism, (2) polarity, and (3) aberrant proliferation inhibition in malignant cells [17], indicating the role of LKB1 as a tumor suppressor gene [18]. Therefore, LKB1 loss promotes cancer evolution and is considered a negative factor in cancer patients [19]. The results of the present investigation indicate, that stage II-III CC patients with loss of LKB1 protein expression exhibited significantly lower DFS (p=0.021) and lower OS (p=0.002) compared to those with LKB1 positive tumors. Thus, the principal finding emerged from the current multivariate analysis, is the prognostic value of LKB1 in adjuvant CRC patients.
It is previously well described, that LKB1 loss confers poor clinical outcome in human gastric cancer, breast cancer and hepatocellular carcinoma [18,20,21]. Furthermore, in 14 eligible studies that met the inclusion criteria, a first comprehensive meta-analysis demonstrated that the decreased LKB1 expression was significantly associated with a poorer OS in solid tumor patients, based on a random effect model [10]. Previous research has demonstrated that LKB1 loss at the transcriptional level, promotes tumor malignancy, not only in lung adenocarcinoma but also in CRC [22]. Another study has revealed that reduced LKB1 expression in patients with gastric cancer is correlated with higher clinical stage, T-stage, lymph-node metastasis and vascular invasion [21]. Similarly, in the present study, a significant correlation of LKB1 loss with certain pathological and clinical parameters of CRC was observed, such as pericolic lymph node involvement (p=0.003) and primary tumors located in the right colon (p=0.032). In contrast, no significant correlation of LKB1 protein expression associated with age, gender and grade was revealed.
The present study failed to demonstrate any statistically significant correlation between LKB1 protein expression and KRAS exon 2 mutation, as has been previously reported in lung adenocarcinomas [23]. On the other hand, our analysis demonstrated significant correlations of LKB1 loss with BRAFV600E mutation and high TS mRNA expression.
In support to previous data that have correlated factors such as KRAS and BRAFV600E mutations, pMMR positive tumors and stage III disease status, with a higher probability of relapse, our results clearly demonstrate that BRAFV600E mutations, pMMR tumors and patients with stage III disease at diagnosis are associated with lower DFS and OS. Although the expression of ERCC1 and TS genes has been shown to be involved in the metabolism of the two main drug categories used in the adjuvant CRC setting, such as oxaliplatin and 5-fluorouracil, respectively [24,25], the current study did not reveal any significant correlation between the ERCC1 and TS mRNA expression and the patients’ outcome. Despite the previously identified prognostic value of NEDD9 and MYC in CC patients [26,27], the current analysis failed to reveal any significant correlation between the NEDD9 and MYC mRNA expression with either DFS or OS in patients with CC receiving adjuvant chemotherapy.
The prognostic or predictive value of LKB1 expression loss in patients with CC is currently unknown. As reported in a previous study, this could be due to the wide range of LKB1 genomic alterations observed in sporadic cancers, emerging the challenge of developing a single assay capable of the detection of all these alterations combined [28]. Moreover, Sanchez-Cespedes [29] reported that many different types of LKB1 somatic mutations in sporadic cancers have been identified including insertions, deletions, nonsense, and frameshift and missense mutations. However, most of the LKB1 genomic alterations can result in either a truncated and therefore inactive form of the protein [28,29] or the complete absence of the protein. To overcome such a limitation, we considered the in-situ IHC assay as a potentially trustworthy, simple, and cost-effective method for evaluating the expression status of LKB1.
Besides, the robust results for LKB1 expression loss and the large patients’ number of the current study, the finding should be interpreted with caution and mainly as hypothesis generated results. One of the main limitations is the lack of validation sets of patients treated or not treated with adjuvant chemotherapy, in order to elucidate the potential prognostic or predictive role of LKB1 expression loss in CC. Consequently, the design of an independent prospective validation trial is one of the future perspectives of our laboratory, where the prognostic power of LKB1 expression loss would be tested and validated prospectively. In summary, the results of the presented study indicate that loss of LKB1 protein expression is clearly associated with poor outcomes of patients with stage III or high-risk stage II CC and merits further evaluation in larger prospective patients’ cohorts.
Electronic Supplementary Material
Supplementary materials are available at Cancer Research and Treatment website (https://www.e-crt.org).
S1 Table.
Sequence of the primers and probes of all reference and target genes
S2 Fig.
LKB1 positive cytoplasmatic expression in normal mucosa (score 1), intense expression in tumor cells (score 3) (×200).
S3 Fig.
(A, B) LKB1 cytoplasmatic expression in tumor cells (×200). (C, D) LKB1 cytoplasmic and nuclear expression in tumor cells (×200). (E, F) Exclusive nuclear expression of LKB1 in tumor cells (E, ×200; F, ×100).
S5 Fig.
LKB1 positive expression in normal crypts of large bowel intestine, negative expression in colon adenocarcinoma (×100).
S6 Fig.
Quantification of β-actin (internal control gene) and TS (gene of interest) mRNA by real-time quantitative reverse transcription polymerase chain reaction (representative image) in the ABI Prism 7900HT. The panel illustrates the logarithmic plot of fluorescence signal during amplification.
References
2. Boland GM, Chang GJ, Haynes AB, Chiang YJ, Chagpar R, Xing Y, et al. Association between adherence to National Comprehensive Cancer Network treatment guidelines and improved survival in patients with colon cancer. Cancer. 2013; 119:1593–601.

3. Schmoll HJ, Twelves C, Sun W, O'Connell MJ, Cartwright T, McKenna E, et al. Effect of adjuvant capecitabine or fluorouracil, with or without oxaliplatin, on survival outcomes in stage III colon cancer and the effect of oxaliplatin on postrelapse survival: a pooled analysis of individual patient data from four randomised controlled trials. Lancet Oncol. 2014; 15:1481–92.

4. Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015; 372:2509–20.
6. Forster LF, Defres S, Goudie DR, Baty DU, Carey FA. An investigation of the Peutz-Jeghers gene (LKB1) in sporadic breast and colon cancers. J Clin Pathol. 2000; 53:791–3.

8. Lee SM, Choi JE, Na YK, Lee EJ, Lee WK, Choi YY, et al. Genetic and epigenetic alterations of the LKB1 gene and their associations with mutations in TP53 and EGFR pathway genes in Korean non-small cell lung cancers. Lung Cancer. 2013; 81:194–9.

9. Carretero J, Medina PP, Pio R, Montuenga LM, Sanchez-Cespedes M. Novel and natural knockout lung cancer cell lines for the LKB1/STK11 tumor suppressor gene. Oncogene. 2004; 23:4037–40.

10. Xiao J, Zou Y, Chen X, Gao Y, Xie M, Lu X, et al. The prognostic value of decreased LKB1 in solid tumors: a meta-analysis. PLoS One. 2016; 11:e0152674.

11. Souglakos J, Philips J, Wang R, Marwah S, Silver M, Tzardi M, et al. Prognostic and predictive value of common mutations for treatment response and survival in patients with metastatic colorectal cancer. Br J Cancer. 2009; 101:465–72.

12. Benlloch S, Paya A, Alenda C, Bessa X, Andreu M, Jover R, et al. Detection of BRAF V600E mutation in colorectal cancer: comparison of automatic sequencing and real-time chemistry methodology. J Mol Diagn. 2006; 8:540–3.
13. Saridaki Z, Tzardi M, Papadaki C, Sfakianaki M, Pega F, Kalikaki A, et al. Impact of KRAS, BRAF, PIK3CA mutations, PTEN, AREG, EREG expression and skin rash in ≥2 line cetuximab-based therapy of colorectal cancer patients. PLoS One. 2011; 6:e15980.
14. Cushman-Vokoun AM, Stover DG, Zhao Z, Koehler EA, Berlin JD, Vnencak-Jones CL. Clinical utility of KRAS and BRAF mutations in a cohort of patients with colorectal neoplasms submitted for microsatellite instability testing. Clin Colorectal Cancer. 2013; 12:168–78.

15. Papadaki C, Mavroudis D, Trypaki M, Koutsopoulos A, Stathopoulos E, Hatzidaki D, et al. Tumoral expression of TXR1 and TSP1 predicts overall survival of patients with lung adenocarcinoma treated with first-line docetaxel-gemcitabine regimen. Clin Cancer Res. 2009; 15:3827–33.

16. Rowan A, Churchman M, Jefferey R, Hanby A, Poulsom R, Tomlinson I. In situ analysis of LKB1/STK11 mRNA expression in human normal tissues and tumours. J Pathol. 2000; 192:203–6.
17. Gan RY, Li HB. Recent progress on liver kinase B1 (LKB1): expression, regulation, downstream signaling and cancer suppressive function. Int J Mol Sci. 2014; 15:16698–718.
18. Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009; 9:563–75.

19. Yang JY, Jiang SH, Liu DJ, Yang XM, Huo YM, Li J, et al. Decreased LKB1 predicts poor prognosis in pancreatic ductal adenocarcinoma. Sci Rep. 2015; 5:10575.

20. Shen Z, Wen XF, Lan F, Shen ZZ, Shao ZM. The tumor suppressor gene LKB1 is associated with prognosis in human breast carcinoma. Clin Cancer Res. 2002; 8:2085–90.
21. Sun J, Ling B, Xu X, Ma R, Li G, Cao X, et al. Decreased expression of tumor-suppressor gene LKB1 correlates with poor prognosis in human gastric cancer. Anticancer Res. 2016; 36:869–75.
22. He TY, Tsai LH, Huang CC, Chou MC, Lee H. LKB1 loss at transcriptional level promotes tumor malignancy and poor patient outcomes in colorectal cancer. Ann Surg Oncol. 2014; 21 Suppl 4:S703–10.

23. Koivunen JP, Kim J, Lee J, Rogers AM, Park JO, Zhao X, et al. Mutations in the LKB1 tumour suppressor are frequently detected in tumours from Caucasian but not Asian lung cancer patients. Br J Cancer. 2008; 99:245–52.

24. Donada M, Bonin S, Nardon E, De Pellegrin A, Decorti G, Stanta G. Thymidilate synthase expression predicts longer survival in patients with stage II colon cancer treated with 5-flurouracil independently of microsatellite instability. J Cancer Res Clin Oncol. 2011; 137:201–10.

25. Liang J, Jiang T, Yao RY, Liu ZM, Lv HY, Qi WW. The combination of ERCC1 and XRCC1 gene polymorphisms better predicts clinical outcome to oxaliplatin-based chemotherapy in metastatic colorectal cancer. Cancer Chemother Pharmacol. 2010; 66:493–500.

26. Lee KS, Kwak Y, Nam KH, Kim DW, Kang SB, Choe G, et al. Favorable prognosis in colorectal cancer patients with coexpression of c-MYC and ss-catenin. BMC Cancer. 2016; 16:730.

27. Li P, Zhou H, Zhu X, Ma G, Liu C, Lin B, et al. High expression of NEDD9 predicts adverse outcomes of colorectal cancer patients. Int J Clin Exp Pathol. 2014; 7:2565–70.
Fig. 1.
Disease-free survival according to LKB1 protein expression loss by immunohistochemistry. DFS, disease-free survival; HR, hazard ratio; CI, confidence interval.
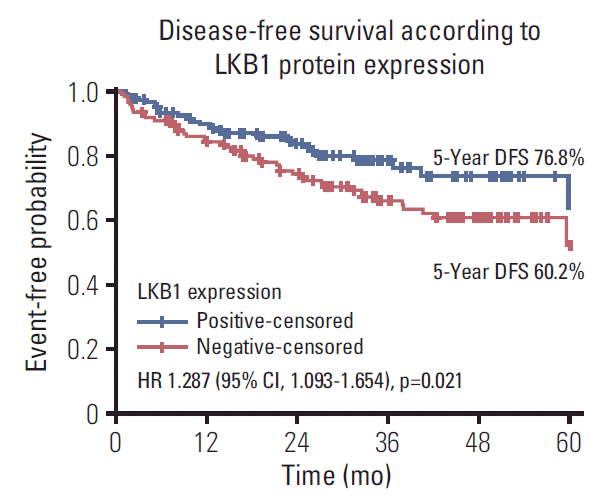
Fig. 2.
Overall survival according to LKB1 protein expression loss by immunohistochemistry. HR, hazard ratio; CI, confidence interval.
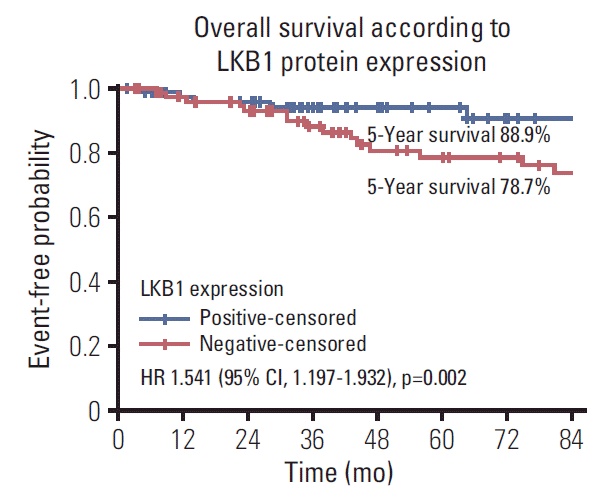
Table 1.
Stage II or III patients: clinical characteristics and pathological features
Table 2.
Laboratory analysis
Table 3.
Correlation of LKB1 expression with patients’ characteristics and tumor’s features and DNA markers
| LKB1 protein expression (n=254) | Negative (n=117) | Positive (n=137) | p-value |
|---|---|---|---|
| Age, median (min-max, yr) | 65 (33-75) | 67 (37-75) | 0.147a) |
| Age group (yr) | |||
| ≤ 70 | 72 (61.5) | 86 (62.8) | 0.897b) |
| > 70 | 45 (38.5) | 51 (37.2) | |
| Sex | |||
| Male | 72 (61.5) | 74 (54.0) | 0.253c) |
| Female | 45 (38.5) | 63 (46.0) | |
| Lymph node status | |||
| N0 | 33 (28.2) | 68 (49.6) | 0.003b) |
| N1-2 | 84 (71.8) | 69 (50.4) | |
| Tumor location | |||
| Right | 49 (41.9) | 41 (29.9) | 0.032b) |
| Left | 68 (58.1) | 96 (70.1) | |
| Primary tumor (T) | |||
| T2-T3 | 74 (36.8) | 114 (56.7) | 0.008b) |
| T4 | 10 (5.0) | 3 (1.5) | |
| Grade | |||
| Low grade | 70 (59.8) | 90 (65.7) | 0.880b) |
| High grade | 47 (38.5) | 47 (34.3) | |
| BRAFV600E status (n=236) | ND 7 | ND 11 | |
| Wild type (n=223) | 100 (90.9) | 123 (97.6) | 0.024 |
| Mutant (n=13) | 10 (9.1) | 3 (2.4) | |
| KRAS exon 2 (n=246) | ND 6 | ND 5 | |
| Wild type (n=224) | 70 (62.7) | 91 (70.1) | 0.328 |
| Mutant (n=22) | 41 (37.3) | 41 (29.9) | |
| MMR status (n=235) | ND 9 | ND 16 | |
| Proficient | 96 (88.9) | 99 (81.8) | 0.102 |
| Deficient | 12 (11.1) | 22 (18.2) | |
| ERCC1 mRNA expression (n=246) | ND 4 | ND 4 | |
| High | 59 (52.2) | 69 (51.9) | 0.609 |
| Low | 54 (47.8) | 64 (48.1) | |
| MYC mRNA expression (n=249) | ND 2 | ND 3 | |
| High | 55 (47.8) | 69 (51.5) | 0.612 |
| Low | 60 (52.2) | 65 (48.5) | |
| NEDD9 mRNA expression (n=247) | ND 3 | ND 4 | |
| High | 58 (50.9) | 64 (48.1) | 0.703 |
| Low | 56 (49.1) | 69 (51.9) | |
| TS mRNA expression (n=226) | ND 15 | ND 13 | |
| High | 58 (56.9) | 49 (39.5) | 0.041 |
| Low | 44 (43.1) | 75 (60.5) |
Table 4.
Univariate analysis for median disease-free and overall survival
Table 5.
Multivariate analysis for median disease-free and overall survival




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download