Abstract
Purpose
MicroRNAs (miRNAs) are a group of small non-coding RNAs involved in different cancers, including lung cancer. Here, we aim to investigate the expression profiles of circulating miRNAs and their roles contributed to the progress of lung cancer.
Materials and Methods
The levels of circulating miRNA in lung cancer patients were investigated by miRNAs assay. Then we predicted the target genes of aberrantly expressing miRNAs by searching genetic databases. Based on the A549 cells transfected with miR-1246 mimics or miR-1246 inhibitor,we further measured the roles of miR-1246 involving in the epithelial-mesenchymal transition (EMT), migration and invasion capacities of lung cancer cells in vitro. Finally, we detected the effects of miR-1246 on glycogen synthase kinase-3β (GSK-3β)/β-catenin pathway by immunofluorescence and Western blot, respectively.
Results
We identified that 14 miRNAs were aberrantly expressed in the serum of lung cancer patients. Among them, miR-1246 was the most up-regulated. The cell assays indicated that miR-1246 significantly increased the migration and invasion capabilities of A549 lung cancer cells. Meanwhile, immunofluorescence analysis revealed that miR-1246 promoted EMT process of A549 cells accompanying with decreasing E-cadherin expression, while increasing vimentin and transforming growth factor β (TGF-β) expression. Furthermore, an online tool predicated that miR-1246 might bind to 3′-untranslated region of GSK-3β, which was confirmed by overexpression and knockdown of miR-1246 assays.
Lung cancer is the leading cause of cancer-related death in the world. About 1.8 million people are diagnosed with lung cancer each year, and the 5-year survival rate ranges about 4%-17% depending on stage and regional differences [1-3]. Despite rapid progresses have been made in the field of diagnostic technology and therapeutic methods, most of lung cancer patients are still diagnosed at advanced stages with poor prognosis. To date, there are limited drugs and therapeutic interventions for the majority of patients with lung cancer. Metastasis is the main factor that results in the poor prognosis of lung cancer patients [4,5]. Thus, there is an urgent requirement to identify novel biomarkers for early diagnosis and explore therapeutic strategies that can specifically alleviate tumor burden, especially reduce metastasis, prolong survival of lung cancer patients [5-8].
Epithelial-mesenchymal transition (EMT) is a program of transforming polarized epithelial cells into cells with mesenchymal phenotypes and functions, such as losing the ability of cell-cell adhesion and the phenotypes related to epithelial cells, while increasing mesenchymal cell markers and migratory capacity, resisting to apoptosis, producing extracellular matrix components, etc. [9]. It is well known that EMT plays important roles in a variety of biological processes, for instance, embryonic development, organ formation, wound healing, and fibrosis. Recent studies in cancer have revealed that initiation of metastasis requires invasion, which is enabled by EMT, and loss of E-cadherin is considered to be a fundamental event in EMT [10,11].
MicroRNAs (miRNAs) are a group of small non-coding single-stranded RNAs with approximately 20-23 nucleotides. They are transcribed by RNA polymerase II and then cleaved sequentially by Drosha and Dicer to form miRNA. Through binding to 3' untranslated region (3'-UTR) of target mRNA, miRNAs regulate post-transcriptional expression, accounting for multiple physiological processes like proliferation, differentiation, and apoptosis [1,12].
Recently, studies have demonstrated that the miRNAs were aberrantly expressed in tumor tissue and/or blood of patients with different tumors, including lung cancer[3,13,14]. More efforts have focused on miRNA as a crucial regulator involved in Wnt/β-catenin, Notch and Hedgehog pathway, etc. [15,16]. The Wnt signaling pathway has served as a critical regulator in lung development as well as physiological and pathophysiological processes of adult lung [17,18]. β-Catenin is a key protein in Wnt/β-catenin signaling cascades, and accounts for varying activities, including embryonic development, stem cell maintenance, tumorigenesis, and metastasis. Glycogen synthase kinase-3β (GSK-3β) is a negative regulator of Wnt/β-catenin signaling pathway, which locates in the upstream of β-catenin, and inhibits its excessive activation [19]. Otsuki et al. [11] reported that suppressing GSK-3β could activate Wnt/β-catenin pathway, then promote EMT and metastasis in lung cancer. Together, these data show that aberrant expression of specific miRNAs may act as biomarkers for diagnosis, prediction of prognosis, and promising targeted therapeutic agents in lung cancer patients [14,18]. Although there are increasing studies on miRNAs and lung cancer [2,3,20], to our knowledge, the level of circulating miRNAs in clinical patients with lung cancer, and the mechanism of altered circulating miRNAs on the carcinogenesis of lung cancer have not been well demonstrated.
In the present study, we investigated the expression profile of circulating miRNAs in lung cancer patients, and then explored the roles of remarkably altered miR-1246 involving in malignancy of lung cancer cells.
In this study, we recruited 11 primary lung cancer patients (which composed of 4 squamous carcinomas, 5 adenocarcinomas, and 2 small cell lung cancer cases) and five healthy control individuals (not affected by lung diseases or any neoplastic disorder). The peripheral blood were collected from lung cancer patients prior to therapeutic interventions (surgery resection, chemotherapy, or radiotherapy). The final diagnosis of lung cancer was confirmed by pathological examination of surgical tumor resections.
MiRNA expression profiles in the serum of lung cancer patients and matched controls were determined by a high-throughput real-time quantitative polymerase chain reaction (RT-qPCR) after we sent the collected sera to Wcgene Biotech Shanghai Company (Shanghai, China). Briefly, total RNAs were isolated from 250 μL serum sample with 750 μL TRIZOL LS Reagent (Invitrogen, Carlsbad, CA) and 1 μL (20 nM) synthesized internal control 1 miRNA to homogenize samples and incubated for 5 minutes at room temperature. Subsequently, 200 μL chloroform were added for separating RNA. The purified RNAs were polyadenylated through a poly(A) polymerase reaction and then reversed-transcribed into cDNA. Individual miRNAs were quantified by qPCR using gene-specific primers (10 μM) for the 36 miRNAs of interest. Detailed step-by-step protocols for RNA isolation, first-strand cDNA synthesis and qPCR are available in the Supplementary Material 1. The miRNA assay analysis was carried out as details described on the website of Wcgene (http://www.wcgene.com).
Non-small cell lung cancer (NSCLC) cell line A549 (lung adenocarcinoma) was obtained from the China Center for Type Culture Collection (CCTCC) (Wuhan, China). Cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum (FBS) (Invitrogen), 100 U/mL penicillin, and 50 μg/mL streptomycin in an incubator at 37°C under humidified atmosphere containing 5% CO2. For overexpression of miR-1246 assay, cells were transfected with 100 pmoL of miR-1246 mimics (Gene Pharma, Shanghai, China) or negative control (NC) miRNA using Lipofectamine 2000 (Invitrogen, Thermo Scientific) according to the manufacturer’s protocol. For knockdown assay, cells were transfected with miR-1246 inhibitor or inhibitor negative control (Inh-NC).
A549 cells were transfected with miR-1246 mimics or NC, inhibitor, or Inh-NC for 48 hours. In the migration assay, 2×104 cells suspend in 200 μL serum-free medium were added to the upper chamber of a Boyden transwell with 8 μm pores (Costar #3422, Corning Incorporated, Corning, NY). Subsequently, 500 μL medium with 10% FBS was filled into the lower chamber. After incubation for 24 hours at 37°C in a 5% CO2 humidified incubator, cells that did not migrate through the pores and remained on the surface of upper chamber membrane were gently removed by a wet cotton wool. The cells invading the filters were fixed with 4% formaldehyde (Beyotime Biotechnology, Shanghai, China), and then stained with 0.1% crystal violet solution (Beyotime Biotechnology) for 30 minutes. For the invasion assay, the upper chamber of transwell insert was first coated with Matrigel Matrix (BD Biosciences, Franklin Lakes, NJ). Aside from incubating for 48 hours, the other procedures were similar to those in migration assay. Images were captured with light microscope (Olympus, Tokyo, Japan) and cell numbers were counted in five randomly selected fields of each insert.
RT-qPCR assay was applied to evaluate the expression level of mRNA related to the EMT program in A549 cells. Total RNA was isolated from A549 cells with Trizol reagent (Gibco, Rockville, MD), the concentration and purity of isolated RNA were assessed at 260/280 nm by a nanodrop spectrophotometer (ND-2000, Thermo Fisher Scientific, Waltham, MA). For cDNA synthesis, 1 μg RNA was reversely transcribed in a 10 μL reaction mixture, containing 1 μL PrimeScript RT enzyme mix, according to manufacturer’s instructions (TaKaRa, Otsu, Japan). Real-time PCR was conducted in 20 μL reaction system including 1 μL of cDNA, 10 μL SYBR Premix Ex Taq II (TaKaRa), 0.2 μL (10 μM) of both forward and reverse primers. The PCR cycles were as follows: 95°C/30 seconds, 40 cycles of 95°C/5 seconds, 60°C/30 seconds, and the melt-curve analysis was carried out at the end of each experiment to determine that a single product per primer pair was amplified. The amplification and analysis were performed using a StepOne Software v2.1 (ABI, Foster City, CA). All qRT-PCR reactions were run in triplicate and the relative expression levels of specific gene mRNA were calculated by using the comparative threshold method (2−ΔΔCt). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as housekeeping gene. The primer sequences used were as follows: E-cadherin primers, forward 5′-CTGCTCTTGCTGTTTCTTCGG-3′, reverse 5′-AAGTCAAAGTCCTGGTCCTCTTCTC-3′; vimentin primers, forward 5′-CACCTTCGTGAATACCAAGACCTG-3′, reverse 5′-GAAATCCTGCTCTCCTCGCC-3′; transforming growth factor β (TGF-β) primers, forward 5′-CGCCGCCTACTACTGTGAGG-3′, reverse 5′-GATGAA-GTGGACCAGCGTCTG-3′; GAPDH primers, forward 5′-GGTAGTCTCCTAAGACTTCAA-3′, reverse 5′-CCACCCTGTTGCTGTAGCC-3′.
A549 cells were cultured on the glass cover slips and then transfected as the above stated. After incubation, cells were fixed with 4% formaldehyde for 15 minutes at 4°C, permeabilized with 0.01% Triton X-100 for 5 minutes, and blocked with 1% bovine serum albumin for 30 minutes at room temperature. Then the prepared cells were incubated with the primary antibody GSK-3β (#12456, 1:100, Cell Signaling Technology, Danvers, MA), or β-catenin (#8814, 1:100, Cell Signaling Technology) for 24 hours at 4°C. In the next day, the cells were stained with isotype-specific secondary antibody (Alexa Fluor 594-AffiniPure donkey anti-rabbit IgG) (Jackson ImmunoResearch, West Grove, PA) for 1 hour. DAPI was used to label the nuclei. Following staining, background autofluorescence was quenched by bathing slides in 0.1% Sudan Black B Solution. The slides were mounted using Prolong Gold (Invitrogen). Sections were visualized with a A1R+ confocal microscope (Nikon Corporation, Tokyo, Japan).
Cells were lysed in RIPA buffer with complete protease inhibitor cocktail (Sigma, St. Louis, MO). The protein was quantified using a BCA Protein Assay Kit (Shanghai ExCell Biology, Inc., Shanghai, China). A total of 50 μg protein per sample was subjected to 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and then transferred onto polyvinylidene difluoride membranes (Millipore, Billerica, MA). After blocked in 5% non-fat milk in TBST (pH 7.4) for 1 hour at room temperature, each membrane was incubated with primary antibodies GSK-3β (#12456, 1:1,000, Cell Signaling Technology), β-catenin (#8814, 1:1,000, Cell Signaling Technology), and β-actin (#4970L, 1:5,000, Cell Signaling Technology) overnight at 4°C. Following incubation with the specific secondary antibodies (peroxidase-conjugated AffiniPure goat anti-rabbit lgG, 1:3,000, Wuhan KeRui Biotechnology, Wuhan, China) for 1 hour, chemiluminescence signal was detected using an electrochemiluminescence kit (Cat. G2014-1, Wuhan Servicebio Technology Co., Ltd.). The density of the immunoreactive bands was analyzed using a KODAK MI software system.
All data derived from at least three independent experiments were presented as mean±standard error of the mean and performed using SPSS ver. 19.0 statistical software (SPSS Inc., Chicago, IL). The p-values were analyzed by one-way ANOVA and Student’s t test. In all analyses, p < 0.05 was considered to be statistically significant.
MiRNA assay was carried out to identify the expression profiles of miRNA in the serum of lung cancer patients. The results revealed that 10 miRNAs were up-regulated (miRNA-1246, miRNA-376a-5p, miRNA-1299, miRNA-373, miRNA-21, miRNA-140-5p, miRNA-432, miRNA-520d-3p, miRNA-211, and miRNA-132-5p) and four miRNAs were down-regulated (miRNA-302b, miRNA-190b, miRNA-5p, and miRNA-505) in lung cancer patients compared to healthy control subjects (Fig. 1). The whole results of miRNA assay are available in the Supplementary Material 2. Among them, miR-1246 was the most obviously expressing alteration miRNA in the lung cancer patients. Then, we predicted the target genes of these aberrantly expressing miRNAs by searching three different miRNAs databases (http://www.targetscan.org, http://www.microrna.org, http://www.microbase.org). It indicated that these miRNAs were involved in various of biological activities (Table 1), strikingly, most of them were related to metastasis. These data suggested that circulating miRNAs in patients with lung cancer may account for tumor progression, especially for metastasis.
According to the above screening results, we came to focus on activities of the highest expressing miR-1246. The abilities of cell migration and invasion are important aspects of cancer metastasis. Transwell assay is widely used to measure these activities. As shown in our study, miR-1246 mimics treatment obviously increased the migration (the left of Fig. 2A, p < 0.05) and invasion (the left of Fig. 2B, p < 0.05) of lung cancer cells. While miR-1246 inhibitor significantly abrogated these abilities compared to Inh-NC transfected group (the right of Fig. 2A and B). Together the above results demonstrated that miR-1246 promoted metastasis of lung cancer cells by increasing the migration and invasion.
In order to investigate the effects of miR-1246 on EMT of lung cancer cells, the expression of mRNAs related to EMT process were assessed by qRT-PCR assay. Our data depicted that the level of E-cadherin, as an epithelial marker, was decreased in A549 cells after transfected with miR-1246 mimics, whereas the expression of mesenchymal marker vimentin, and TGF-β both were increased (p < 0.05) (Fig. 3A). Conversely, A549 cells transfected with miR-1246 inhibitor resulted in an increased expression of E-cadherin, while decreased expressions of vimentin and TGF-β (Fig. 3B). Consistently, both overexpression and knockdown experiments revealed that miR-1246 promoted EMT in A549 cells accompanying with the alteration of E-cadherin, Vimentin, and TGF-β protein expression, respectively. Which indicated that miR-1246 could work as a promoter of EMT in lung cancer metastasis in vitro.
MiRNA is well-known to regulate the activities of target genes, accounting for the behaviors of cell. Given the up-regulation of miR-1246 in serum of patients with lung cancer, we then identified the miRNA gene loci to explore the underlying molecular mechanisms of the miR-1246 impacting on malignancy of lung cancer. By using an online target prediction algorithm mirtarbase (http://mirtarbase.mbc.nctu.edu.tw/php/search.php?opt=search_box&kw=miR-1246&sort=id&order=asc&page=2), we predicted the targets of miR-1246. Among the identified potential targets, we chose to focus on the roles of GSK-3β. First, GSK-3β/β-catenin pathway was previously known as regulating tumor progression [2]. Second, the complementary sequence of miR-1246 was found in the 3′-UTR of GSK-3β mRNA (Fig. 4A) (http://mirtarbase.mbc.nctu.edu.tw/php/detail.php?mirtid=MIRT733922), it depicted that miR-1246 could directly target the 3′-UTR of GSK-3β. In order to verify this prediction, we analyzed the roles of miR-1246 on the expression of GSK-3β by immunofluorescence and western blot assay. As shown in Fig. 4B, D, and E, overexpression of miR-1246 significantly decreased GSK-3β expression (the left of Fig. 4B and D), whereas miR-1246 inhibitor increased GSK-3β expression (the right of Fig. 4B and E), which is a negative regulator of Wnt/β-catenin signaling pathway and locates in the upstream of β-catenin. Taken together, our data suggested that miR-1246 could directly target the 3′-UTR of GSK-3β mRNA, the effect may be as an upstream target of β-catenin in regulating cell activities of lung cancer.
Previous studies have demonstrated that β-catenin, a crucial component of Wnt/β-catenin signaling pathway, can enter into the nucleus and interact with transcription factors to regulate the expression of target gene [17]. Since Wnt/β-catenin signaling pathway is an important regulator for physiological and pathophysiological processes of adult lung, we evaluated the effect of miR-1246 on the nuclear translocation of β-catenin in A549 cells by overexpression and knockdown assay, respectively. The data from overexpression assay indicated that miR-1246 mimics could strongly elevate the level of β-catenin protein in A549 cells (the left of Fig. 4C and D). Meanwhile, miR-1246 inhibitors significantly reduced the β-catenin protein levels compared to the Inh-NC in the knockdown assay (the right of Fig. 4C and E) (p < 0.05). Therefore, miR-1246 could enhance the level of β-catenin in A549 cell.
Taken together, the above findings indicate that miR-1246 could target GSK-3β/β-catenin to regulate Wnt/β-catenin signaling pathway accounting for the progress of lung cancer cells.
Lung cancer remains the leading cause of cancer lethal worldwide partly for lacking early diagnostic markers, and common metastasis, which is the main cause of lung cancer patients’ death [4]. MiRNAs are secreted by cells into body liquids with stable and constant levels, like in peripheral blood (serum and plasma). Since collecting peripheral blood is more easily and less invasive than obtaining the other types of tissue samples, secreted miRNAs are considered to be potential non-invasive biomarkers for diagnosing and tracking disease progression [12,13,21]. Accumulating evidences indicate that miRNA expression profiles in the serum of patient represents molecular signatures, which is not only accounting for tumorigenesis, but also involving in tumor invasion and metastasis [22,23]. However, there is limited literature concerning the levels of circulating miRNAs and further exploring their roles in the progression of lung cancer. After using a high-through miRNA assay on a small number of serum samples, we found that 14 circulating miRNAs were aberrantly expressed in lung cancer patients. Among them, miR-1246 was the most remarkably altered miRNAs in the serum of lung cancer patients. Similarly, Zhang et al. [24] identified that miR-1246 was enriched in CD166+ lung tumor-initiating cell (TIC, also referred as cancer stem cell [CSC]) from solid tumors relative to CD166– non-TIC in lung cancer patients. Kim et al. [25] discovered that anti-miR-1246 reduced the expressions of stemness-related and EMT-related markers which associated with characteristics of CSCs, such as cellular proliferation, tumorigenesis, colony formation, and invasiveness, etc. in NSCLC cell lines in vitro. Yuan’s group reported that extracellular miR-1246, which was one of different bio-fluids, promoted lung cancer cell proliferation and enhanced radioresistance [26]. Thus, our data imply miR-1246 may be used as a serum biomarker for lung cancer patients.
The EMT process is closely related to initiate the metastasis in many cancer patients [9,10]. Importantly, miRNAs have been shown to regulate EMT [20,25]. Therefore, in this study, we investigated whether miR-1246 alteration accounted for EMT. Here, we found that miR-1246 expression was closely related to the EMT process of lung cancer cells by gaining mesenchymal markers and losing epithelial markers. MiR-1246 could significantly decrease E-cadherin expression and increase vimentin and TGF-β expression, meanwhile, knockdown miR-1246 notably inhibited the EMT. Interestingly, the data from predicting downstream genes of those aberrantly expressed miRNAs also indicated that major of miR-1246’s downstream targets were related to EMT process (Table 1).
It is well known that miRNAs regulate target genes to perform various biological and pathological function [1]. Both GSK-3β and β-catenin are crucial regulators of Wnt/β-catenin signaling implicated in the progress of various tumors [2,17]. Since miR-1246 enhanced lung cancer cell migration and invasion (Fig. 2), we further investigated whether miR-1246 impacted on the expression of GSK-3β and β-catenin during lung cancer cell metastasis. The results derived from overexpression and knockdown assays show that miR-1246 could target the expressions of GSK-3β and β-catenin. These findings are consistent with the prior reports that miR-26a enhanced invasiveness by suppressing GSK-3β in lung cancer and octamer 4/miR-1246 signaling axis promoted Wnt/β-catenin activation in liver CSCs [2,27]. Which indicates that miR-1246 can act as a promoter of lung tumor cells metastasis partly due to down-regulating GSK-3β while up-regulating β-catenin.
Together, we provide strong evidence that miR-1246 may be exploited for the early diagnosis of lung cancer. Moreover, we demonstrated that miR-1246 enhanced EMT activity of A549 cells, a crucial process for metastasis, supporting that miR-1246 can act as a promoter of A549 metastasis. We also dissected the underlying molecular mechanism of miR-1246 involved in regulating Wnt/β-catenin signaling pathway, by directly targeting the expression of GSK-3β and β-catenin, the effect may be partly contributing to the invasion and metastasis of lung cancer. In conclusion, our data illustrated that miR-1246 may behave as a non-invasive biomarker and therapeutic target of lung cancer.
Electronic Supplementary Material
Supplementary materials are available at Cancer Research and Treatment website (https://www.e-crt.org).
ACKNOWLEDGMENTS
This work was supported by Natural Science Foundation of Hubei Province (2017CFB573), Scientific Research and Development Projects of Yichang Municipality (A16-301-29, A17-301-24) and Open Foundation of Hubei Key Laboratory of Tumor Microenvironment and Immunotherapy (Three Gorges University) (2015KZL-13, 2018KZL06).
References
1. Uddin A, Chakraborty S. Role of miRNAs in lung cancer. J Cell Physiol. 2018; Apr. 20. [Epub]. https://doi.org/10.1002/jcp.26607.

2. Lin G, Liu B, Meng Z, Liu Y, Li X, Wu X, et al. MiR-26a enhances invasive capacity by suppressing GSK3beta in human lung cancer cells. Exp Cell Res. 2017; 352:364–74.
3. Jiang W, Wei K, Pan C, Li H, Cao J, Han X, et al. MicroRNA-1258 suppresses tumour progression via GRB2/Ras/Erk pathway in non-small-cell lung cancer. Cell Prolif. 2018; 51:e12502.

4. Wu KL, Tsai MJ, Yang CJ, Chang WA, Hung JY, Yen CJ, et al. Liver metastasis predicts poorer prognosis in stage IV lung adenocarcinoma patients receiving first-line gefitinib. Lung Cancer. 2015; 88:187–94.

5. Reck M, Popat S, Reinmuth N, De Ruysscher D, Kerr KM, Peters S, et al. Metastatic non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014; 25 Suppl 3:iii27–39.

6. Bica-Pop C, Cojocneanu-Petric R, Magdo L, Raduly L, Gulei D, Berindan-Neagoe I. Overview upon miR-21 in lung cancer: focus on NSCLC. Cell Mol Life Sci. 2018; 75:3539–51.

7. Song L, Peng L, Hua S, Li X, Ma L, Jie J, et al. miR-144-5p enhances the radiosensitivity of non-small-cell lung cancer cells via targeting ATF2. Biomed Res Int. 2018; 2018:5109497.

8. Kim S, Kim TM, Kim DW, Kim S, Kim M, Ahn YO, et al. Acquired resistance of MET-amplified non-small cell lung cancer cells to the MET inhibitor capmatinib. Cancer Res Treat. 2019; 51:951–62.

9. Nowrin K, Sohal SS, Peterson G, Patel R, Walters EH. Epithelial-mesenchymal transition as a fundamental underlying pathogenic process in COPD airways: fibrosis, remodeling and cancer. Expert Rev Respir Med. 2014; 8:547–59.

10. Sung WJ, Kim H, Park KK. The biological role of epithelial-mesenchymal transition in lung cancer (Review). Oncol Rep. 2016; 36:1199–206.

11. Otsuki Y, Saya H, Arima Y. Prospects for new lung cancer treatments that target EMT signaling. Dev Dyn. 2018; 247:462–72.

12. Martinez-Rivera V, Negrete-Garcia MC, Avila-Moreno F, Ortiz-Quintero B. Secreted and tissue miRNAs as diagnosis biomarkers of malignant pleural mesothelioma. Int J Mol Sci. 2018; 19:E195.
13. Moshiri F, Salvi A, Gramantieri L, Sangiovanni A, Guerriero P, De Petro G, et al. Circulating miR-106b-3p, miR-101-3p and miR-1246 as diagnostic biomarkers of hepatocellular carcinoma. Oncotarget. 2018; 9:15350–64.

14. Li L, Sun Y, Feng M, Wang L, Liu J. Clinical significance of blood-based miRNAs as biomarkers of non-small cell lung cancer. Oncol Lett. 2018; 15:8915–25.
15. Yang Y, Sun Y, Wu Y, Tang D, Ding X, Xu W, et al. Downregulation of miR-3127-5p promotes epithelial-mesenchymal transition via FZD4 regulation of Wnt/beta-catenin signaling in non-small-cell lung cancer. Mol Carcinog. 2018; 57:842–53.
16. Baranwal S, Alahari SK. miRNA control of tumor cell invasion and metastasis. Int J Cancer. 2010; 126:1283–90.

17. Skronska-Wasek W, Gosens R, Konigshoff M, Baarsma HA. WNT receptor signalling in lung physiology and pathology. Pharmacol Ther. 2018; 187:150–66.

18. Tanaka K, Kumano K, Ueno H. Intracellular signals of lung cancer cells as possible therapeutic targets. Cancer Sci. 2015; 106:489–96.

19. Krishnamurthy N, Kurzrock R. Targeting the Wnt/beta-catenin pathway in cancer: update on effectors and inhibitors. Cancer Treat Rev. 2018; 62:50–60.

20. Jia Z, Zhang Y, Xu Q, Guo W, Guo A. miR-126 suppresses epithelial-to-mesenchymal transition and metastasis by targeting PI3K/AKT/Snail signaling of lung cancer cells. Oncol Lett. 2018; 15:7369–75.

21. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008; 105:10513–8.

22. Kodahl AR, Lyng MB, Binder H, Cold S, Gravgaard K, Knoop AS, et al. Novel circulating microRNA signature as a potential non-invasive multi-marker test in ER-positive early-stage breast cancer: a case control study. Mol Oncol. 2014; 8:874–83.

23. Ortiz-Quintero B. Cell-free microRNAs in blood and other body fluids, as cancer biomarkers. Cell Prolif. 2016; 49:281–303.

24. Zhang WC, Chin TM, Yang H, Nga ME, Lunny DP, Lim EK, et al. Tumour-initiating cell-specific miR-1246 and miR-1290 expression converge to promote non-small cell lung cancer progression. Nat Commun. 2016; 7:11702.

25. Kim G, An HJ, Lee MJ, Song JY, Jeong JY, Lee JH, et al. HsamiR-1246 and hsa-miR-1290 are associated with stemness and invasiveness of non-small cell lung cancer. Lung Cancer. 2016; 91:15–22.

26. Yuan D, Xu J, Wang J, Pan Y, Fu J, Bai Y, et al. Extracellular miR-1246 promotes lung cancer cell proliferation and enhances radioresistance by directly targeting DR5. Oncotarget. 2016; 7:32707–22.

27. Chai S, Ng KY, Tong M, Lau EY, Lee TK, Chan KW, et al. Octamer 4/microRNA-1246 signaling axis drives Wnt/beta-catenin activation in liver cancer stem cells. Hepatology. 2016; 64:2062–76.
Fig. 1.
Expression profiling of circulating miRNAs in patients with lung cancer. (A) MiRNA assay analysis of circulating miRNAs from the patients with lung cancer was presented in a heat map (11 lung cancer vs. 5 health control; the 11 lung cancer group includes 4 squamous carcinomas, 4 adenocarcinomas, and 2 small cell lung cancer cases). (B) The 14 miRNAs were differentially expressed in 11 lung cancer vs. 5 health control (p < 0.05, fold-change > 2 or < 0.5). A differentially expressed miRNA was considered if their expression levels showed with fold-changes greater than 2.0 or less than 0.5. Red and blue colors indicate increased and decreased expression, respectively.
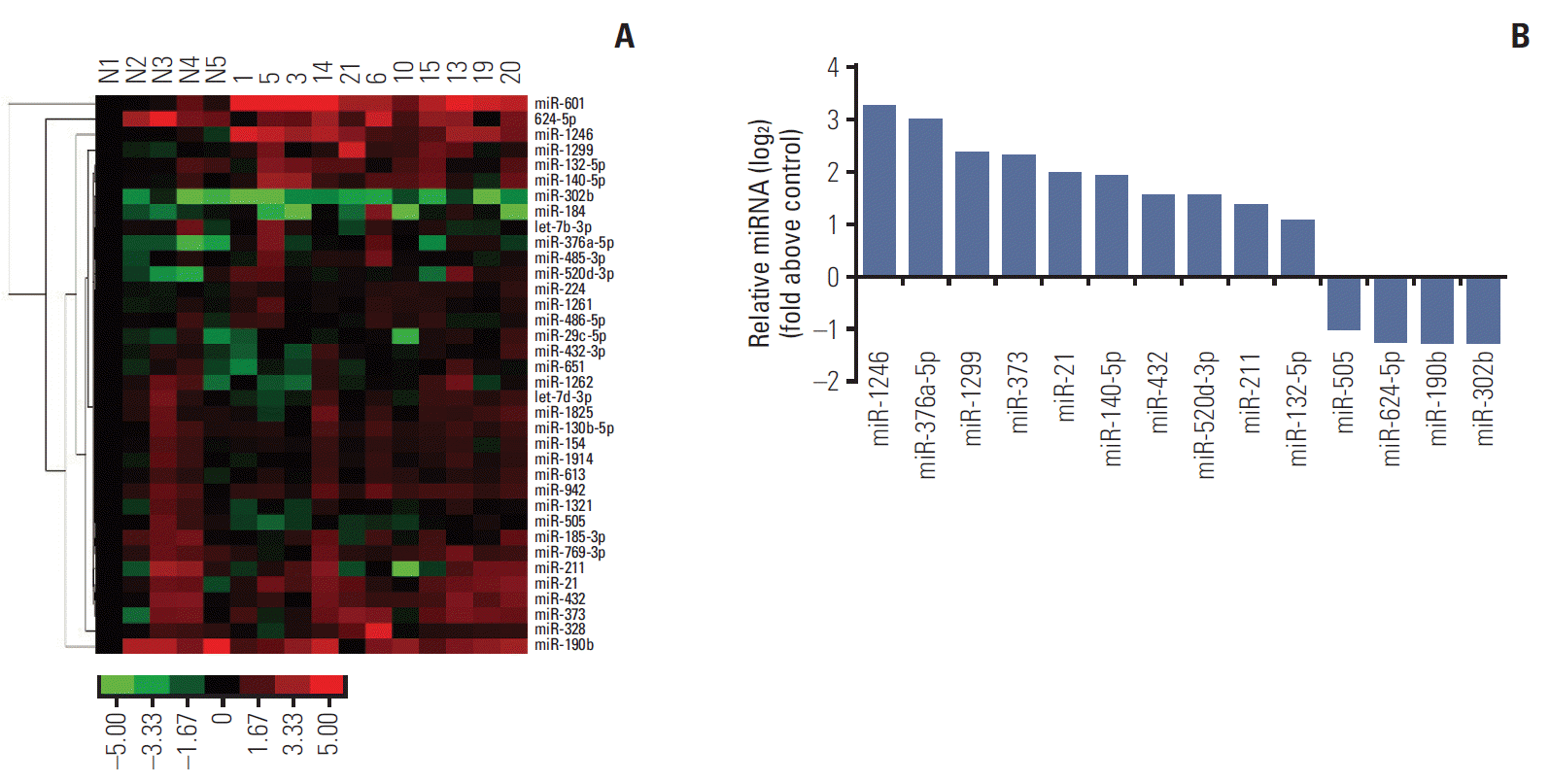
Fig. 2.
MiR-1246 enhances the migration and invasion of A549 cells. Migration and invasion assays were performed after A549 transfected with miR-1246 mimics or negative control (NC) miRNA, miR-1246 inhibitor or inhibitor negative control (Inh-NC) for 48 hours. The cells that migrated and invasive were counted, the representative images were shown. Scale bar=100 μm. (A) The migration ability of A549 cells. (B) The invasion capacity of A549 cells. Experiments were performed at least in triplicate and data are represented as mean±standard error of mean (*p < 0.05).
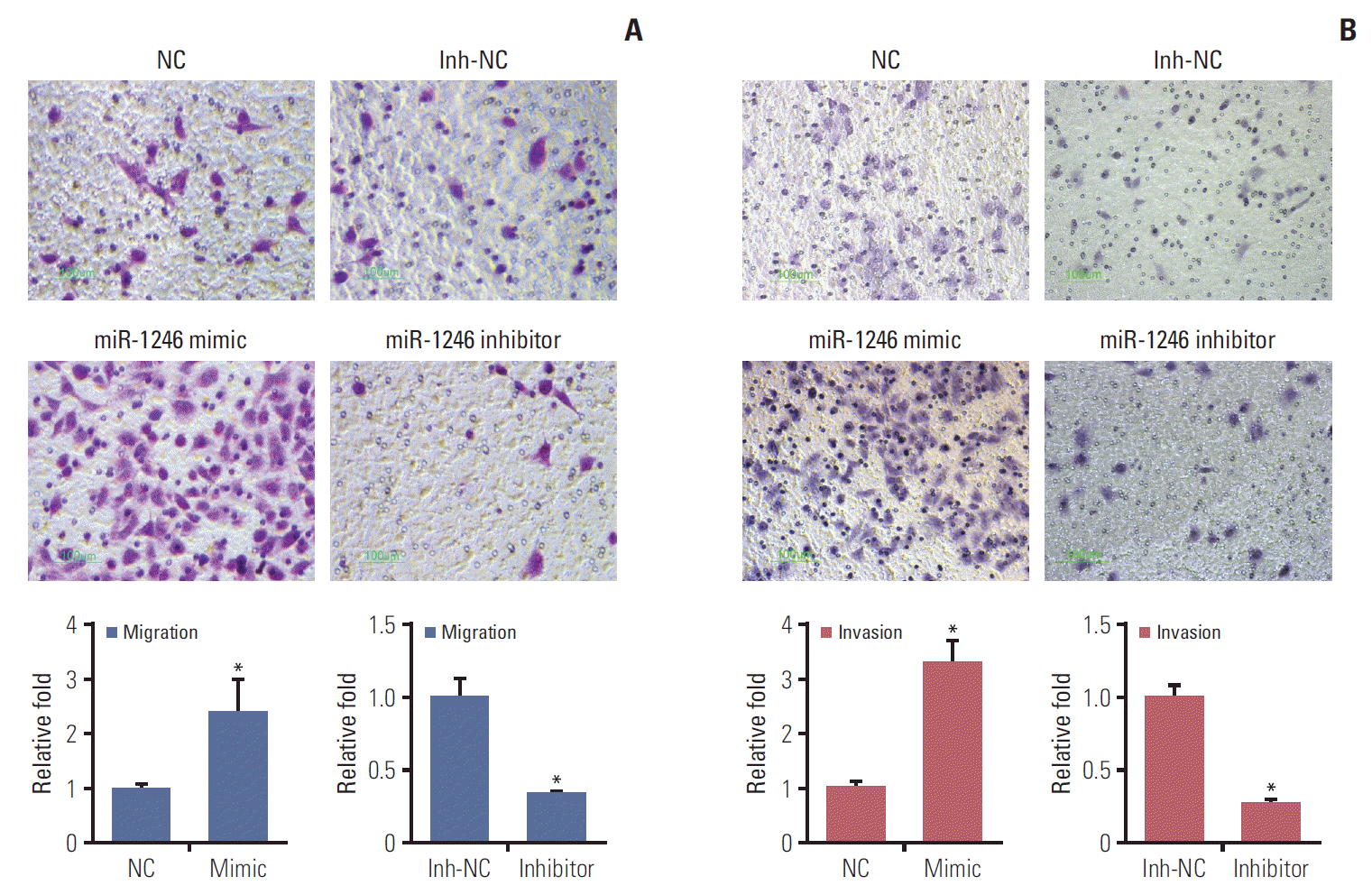
Fig. 3.
MiR-1246 promotes epithelial-mesenchymal transition (EMT) in A549 cells. The mRNA expression of markers (E-cadherin, vimentin, and transforming growth factor β [TGF-β]) related to EMT were measured by real-time quantitative polymerase chain reaction (RT-qPCR) assay. A549 cells were transfected with miR-1246 mimics or negative control (NC), miR-1246 inhibitor, or inhibitor negative control (Inh-NC). After 48 hours, whole cell lysates were prepared and RT-qPCR assay determined the mRNA level of EMT-related markers. (A) The mRNA levels of E-cadherin, vimentin, and TGF-β in A549 cells transfected by miR-1246 mimics or NC. (B) The levels of E-cadherin, vimentin, and TGF-β mRNA in A549 cells transfected with miR-1246 inhibitor or Inh-NC. All experiments were carried out more than three times and results are expressed as mean±standard error of mean (*p < 0.05, **p < 0.01).
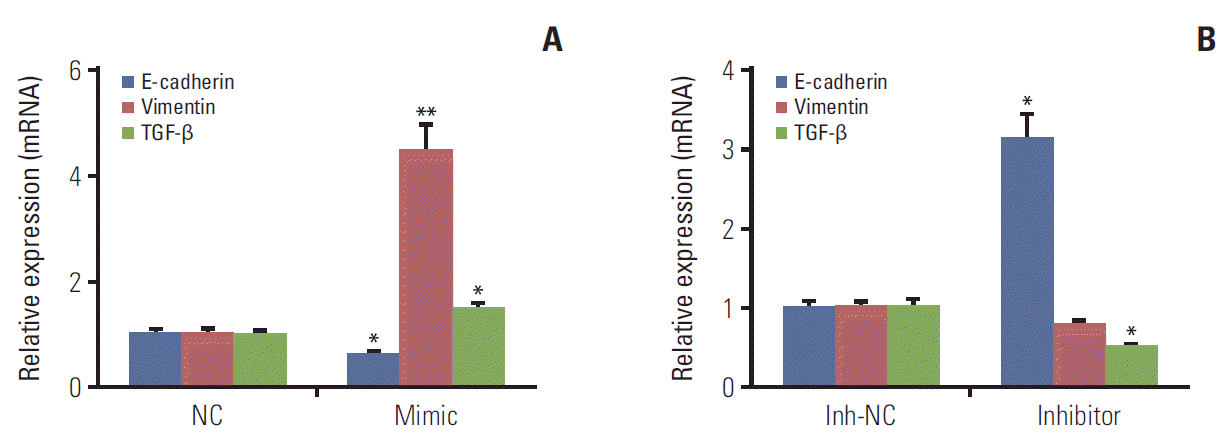
Fig. 4.
MiR-1246 regulates Wnt/β-catenin signaling pathway through targeting glycogen synthase kinase-3β (GSK-3β)/β-catenin. (A) GSK-3β was identified as a putative target of miR-1246 through prediction databases for bioinformatics search. Schematics showing the predicted binding site of miR-1246 in the 3' untranslated region of GSK-3β. (B-E) The protein levels of GSK-3β (B, D, E) and β-catenin (C, D, E) from A549 cells transfected with miR-1246 mimics or negative control (NC), miR-1246 inhibitor, or inhibitor negative control (Inh-NC) were determined by immunofluorescence and western blot assay, respectively. All the values are shown as mean±standard error of mean and pooled from three independent experiments (*p < 0.05).
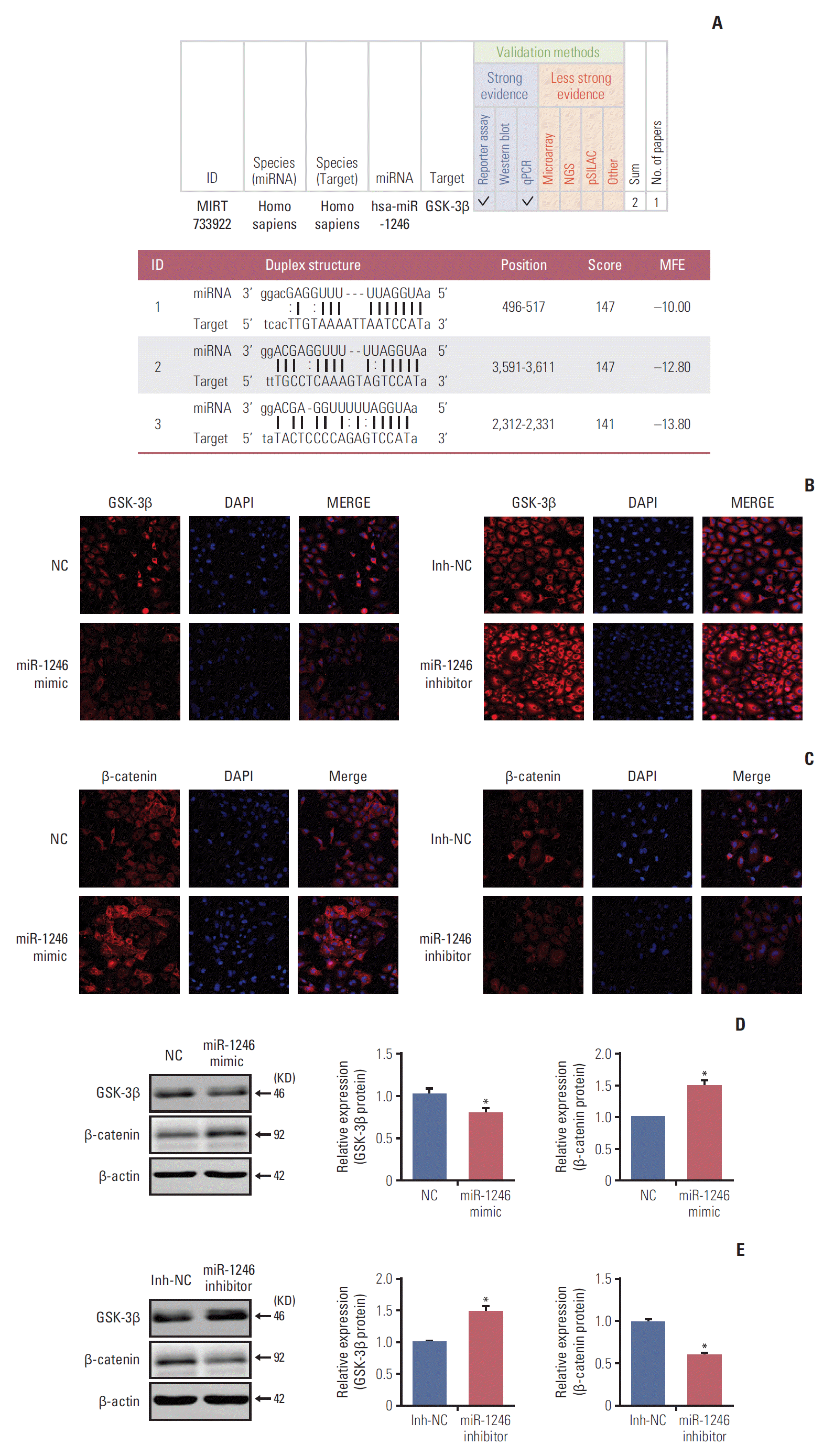
Table 1.
The enriched FunRich pathway of predicted genes




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download