1. Chakravarty D, Tekmal RR, Vadlamudi RK. PELP1: a novel therapeutic target for hormonal cancers. IUBMB Life. 2010; 62:162–9.

2. Girard BJ, Daniel AR, Lange CA, Ostrander JH. PELP1: a review of PELP1 interactions, signaling, and biology. Mol Cell Endocrinol. 2014; 382:642–51.

3. Vadlamudi RK, Manavathi B, Balasenthil S, Nair SS, Yang Z, Sahin AA, et al. Functional implications of altered subcellular localization of PELP1 in breast cancer cells. Cancer Res. 2005; 65:7724–32.

4. Mann M, Zou Y, Chen Y, Brann D, Vadlamudi R. PELP1 oncogenic functions involve alternative splicing via PRMT6. Mol Oncol. 2014; 8:389–400.

5. Rajhans R, Nair S, Holden AH, Kumar R, Tekmal RR, Vadlamudi RK. Oncogenic potential of the nuclear receptor coregulator proline-, glutamic acid-, leucine-rich protein 1/modulator of the nongenomic actions of the estrogen receptor. Cancer Res. 2007; 67:5505–12.

6. Cortez V, Samayoa C, Zamora A, Martinez L, Tekmal RR, Vadlamudi RK. PELP1 overexpression in the mouse mammary gland results in the development of hyperplasia and carcinoma. Cancer Res. 2014; 74:7395–405.

7. Brann DW, Zhang QG, Wang RM, Mahesh VB, Vadlamudi RK. PELP1: a novel estrogen receptor-interacting protein. Mol Cell Endocrinol. 2008; 290:2–7.
8. Kumar R, Zhang H, Holm C, Vadlamudi RK, Landberg G, Rayala SK. Extranuclear coactivator signaling confers insensitivity to tamoxifen. Clin Cancer Res. 2009; 15:4123–30.

9. Habashy HO, Powe DG, Rakha EA, Ball G, Macmillan RD, Green AR, et al. The prognostic significance of PELP1 expression in invasive breast cancer with emphasis on the ER-positive luminal-like subtype. Breast Cancer Res Treat. 2010; 120:603–12.

10. Zhang Y, Dai J, McNamara KM, Bai B, Shi M, Chan MS, et al. Prognostic significance of proline, glutamic acid, leucine rich protein 1 (PELP1) in triple-negative breast cancer: a retrospective study on 129 cases. BMC Cancer. 2015; 15:699.

11. Dang DN, Raj G, Sarode V, Molberg KH, Vadlamudi RK, Peng Y. Significantly increased PELP1 protein expression in primary and metastatic triple-negative breast carcinoma: comparison with GATA3 expression and PELP1's potential role in triple-negative breast carcinoma. Hum Pathol. 2015; 46:1829–35.

12. Vadlamudi RK, Balasenthil S, Broaddus RR, Gustafsson JA, Kumar R. Deregulation of estrogen receptor coactivator proline-, glutamic acid-, and leucine-rich protein-1/modulator of nongenomic activity of estrogen receptor in human endometrial tumors. J Clin Endocrinol Metab. 2004; 89:6130–8.

13. Aust S, Horak P, Pils D, Pils S, Grimm C, Horvat R, et al. The prognostic value of estrogen receptor beta and proline-, glutamic acid- and leucine-rich protein 1 (PELP1) expression in ovarian cancer. BMC Cancer. 2013; 13:115.

14. Nair SS, Guo Z, Mueller JM, Koochekpour S, Qiu Y, Tekmal RR, et al. Proline-, glutamic acid-, and leucine-rich protein-1/modulator of nongenomic activity of estrogen receptor enhances androgen receptor functions through LIM-only coactivator, four-and-a-half LIM-only protein 2. Mol Endocrinol. 2007; 21:613–24.

15. Slowikowski BK, Galecki B, Dyszkiewicz W, Jagodzinski PP. Increased expression of proline-, glutamic acid- and leucine-rich protein PELP1 in non-small cell lung cancer. Biomed Pharmacother. 2015; 73:97–101.
16. Ning Z, Zhang Y, Chen H, Wu J, Song T, Wu Q, et al. PELP1 suppression inhibits colorectal cancer through c-Src downregulation. Oxid Med Cell Longev. 2014; 2014:193523.

17. Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991; 19:403–10.

18. Lakhani SR, Ellis IO, Schnitee SJ, Tan PH, van de Vijver MJ. WHO classification of tumours of the breast. 4th ed. Lyon: IARC Press;2012.
19. Tsang JY, Au WL, Lo KY, Ni YB, Hlaing T, Hu J, et al. PD-L1 expression and tumor infiltrating PD-1+ lymphocytes associated with outcome in HER2+ breast cancer patients. Breast Cancer Res Treat. 2017; 162:19–30.
20. Tsang JY, Ni YB, Chan SK, Shao MM, Law BK, Tan PH, et al. Androgen receptor expression shows distinctive significance in ER positive and negative breast cancers. Ann Surg Oncol. 2014; 21:2218–28.

21. Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thurlimann B, et al. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol. 2013; 24:2206–23.
22. Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004; 10:5367–74.

23. Dimple C, Nair SS, Rajhans R, Pitcheswara PR, Liu J, Balasenthil S, et al. Role of PELP1/MNAR signaling in ovarian tumorigenesis. Cancer Res. 2008; 68:4902–9.

24. Yao T, Wang Q, Zhang W, Bian A, Zhang J. Identification of genes associated with renal cell carcinoma using gene expression profiling analysis. Oncol Lett. 2016; 12:73–8.

25. Grivas PD, Tzelepi V, Sotiropoulou-Bonikou G, Kefalopoulou Z, Papavassiliou AG, Kalofonos H. Expression of ERalpha, ERbeta and co-regulator PELP1/MNAR in colorectal cancer: prognostic significance and clinicopathologic correlations. Cell Oncol. 2009; 31:235–47.
26. Nishimura R, Osako T, Nishiyama Y, Tashima R, Nakano M, Fujisue M, et al. Evaluation of factors related to late recurrence‒later than 10 years after the initial treatment‒in primary breast cancer. Oncology. 2013; 85:100–10.
27. Tang D, Xu S, Zhang Q, Zhao W. The expression and clinical significance of the androgen receptor and E-cadherin in triple-negative breast cancer. Med Oncol. 2012; 29:526–33.

28. Peters AA, Buchanan G, Ricciardelli C, Bianco-Miotto T, Centenera MM, Harris JM, et al. Androgen receptor inhibits estrogen receptor-alpha activity and is prognostic in breast cancer. Cancer Res. 2009; 69:6131–40.
29. Yang L, Ravindranathan P, Ramanan M, Kapur P, Hammes SR, Hsieh JT, et al. Central role for PELP1 in nonandrogenic activation of the androgen receptor in prostate cancer. Mol Endocrinol. 2012; 26:550–61.

30. Nair SS, Mishra SK, Yang Z, Balasenthil S, Kumar R, Vadlamudi RK. Potential role of a novel transcriptional coactivator PELP1 in histone H1 displacement in cancer cells. Cancer Res. 2004; 64:6416–23.

31. Roy S, Chakravarty D, Cortez V, De Mukhopadhyay K, Bandyopadhyay A, Ahn JM, et al. Significance of PELP1 in ER-negative breast cancer metastasis. Mol Cancer Res. 2012; 10:25–33.

32. Rajhans R, Nair HB, Nair SS, Cortez V, Ikuko K, Kirma NB, et al. Modulation of in situ estrogen synthesis by proline-, glutamic acid-, and leucine-rich protein-1: potential estrogen receptor autocrine signaling loop in breast cancer cells. Mol Endocrinol. 2008; 22:649–64.

33. Vadlamudi RK, Kumar R. Functional and biological properties of the nuclear receptor coregulator PELP1/MNAR. Nucl Recept Signal. 2007; 5:e004.

34. Sareddy GR, Vadlamudi RK. PELP1: Structure, biological function and clinical significance. Gene. 2016; 585:128–34.

35. Manavathi B, Nair SS, Wang RA, Kumar R, Vadlamudi RK. Proline-, glutamic acid-, and leucine-rich protein-1 is essential in growth factor regulation of signal transducers and activators of transcription 3 activation. Cancer Res. 2005; 65:5571–7.

36. Truong TH, Hu H, Temiz NA, Hagen KM, Girard BJ, Brady NJ, et al. Cancer stem cell phenotypes in ER(+) breast cancer models are promoted by PELP1/AIB1 complexes. Mol Cancer Res. 2018; 16:707–19.

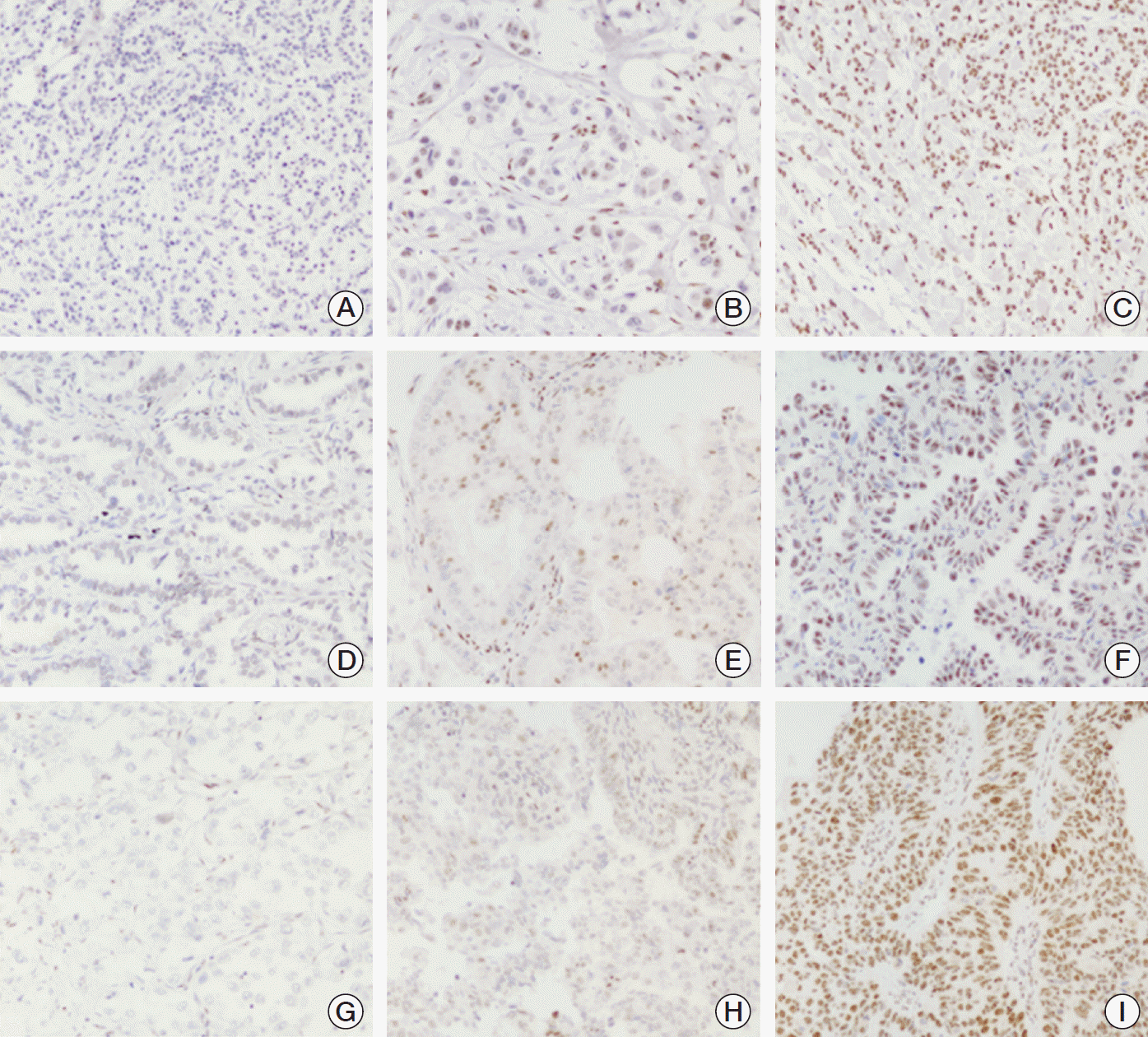
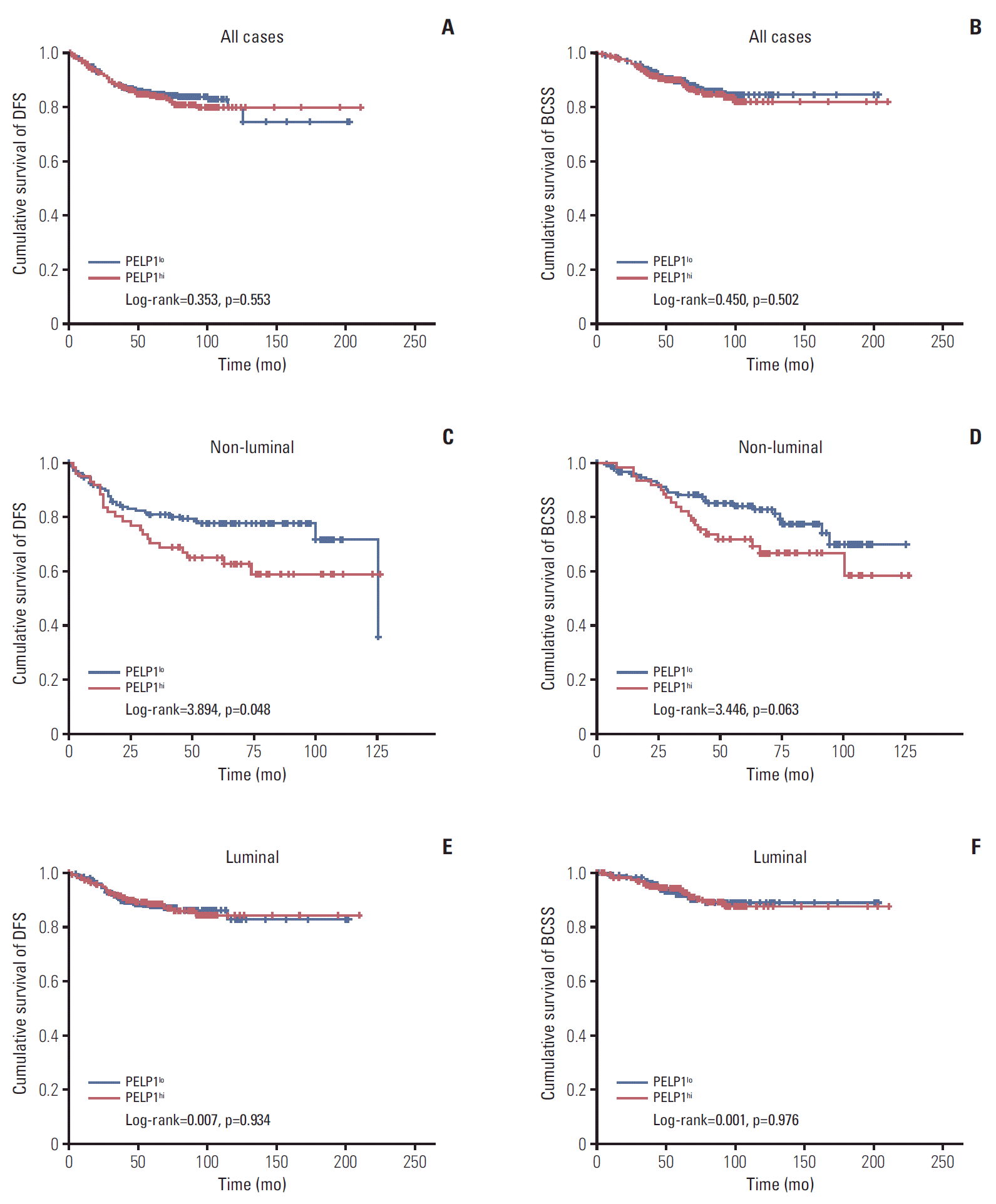
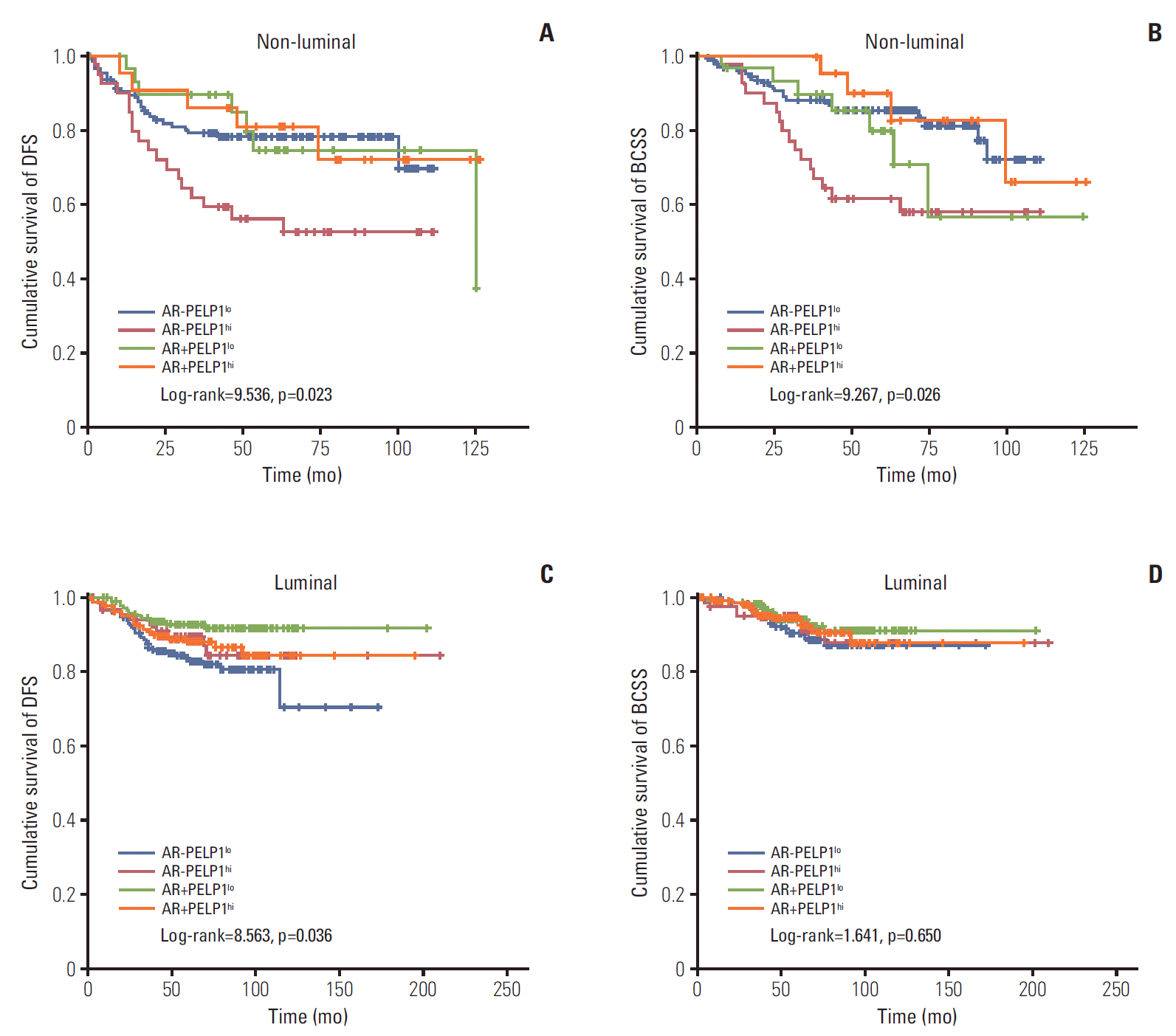




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download