Abstract
Purpose
Half of the world’s gastric cancer cases and the highest gastric cancer mortality rates are observed in Eastern Asia. Although several genome-wide association studies (GWASs) have revealed susceptibility genes associated with gastric cancer, no GWASs have been conducted in the Korean population, which has the highest incidence of gastric cancer.
Materials and Methods
We performed genome scanning of 450 gastric cancer cases and 1,134 controls via Affymetrix Axiom Exome 319 arrays, followed by replication of 803 gastric cancer cases and 3,693 healthy controls.
Results
We showed that the rs2976394 in the prostate stem cell antigen (PSCA) gene is a gastriccancer-susceptibility gene in a Korean population, with genome-wide significance and an odds ratio (OR) of 0.70 (95% confidence interval [CI], 0.64 to 0.77). A strong linkage disequilibrium with rs2294008 was also found, indicating an association with susceptibility. Individuals with the CC genotype of the PSCA gene showed an approximately 2-fold lower risk of gastric cancer compared to those with the TT genotype (OR, 0.47; 95% CI, 0.39 to 0.57). The effect of the PSCA gene on gastric cancer was more prominent in the female population and for diffuse type gastric cancer.
Gastric cancer is the fifth most common type of cancer and the second most common cause of cancer death worldwide [1]. Although the incidence rate has decreased, the absolute number of gastric cancer cases has increased due to population growth and aging [1]. Half of the world’s gastric cancer cases are in Eastern Asia, and the highest mortality rates are also found in this region [2].
Many previous studies have investigated the risk factors for gastric cancer and indicated that Helicobacter pylori infection is one of the most important factors in its etiology [3,4]. However, countries in Eastern Asia, such as Japan and China, have a high gastric cancer incidence rate despite low H. pylori infection rates. In contrast, other countries, such as India, Bangladesh, Pakistan, and Thailand, have a high prevalence of H. pylori infection but low gastric cancer incidence rates, suggesting that individual genetic characteristics may influence the risk of gastric carcinogenesis [5].
In this context, the effects of genetic susceptibility factors for gastric cancer have been explored in several candidate variation approaches, and several recent genome-wide association studies (GWASs) have yielded suggestive variations. Most of the GWASs were conducted in Asian countries, including China and Japan, and the variants associated with gastric cancer were located at the PSCA [6], PLCE1 [7], PTGER4-PKKAA1, ZBTB20 [8], LRFN2, DNAH11 [9], PRKAA1, MUC1, and UNC5CL genes [10]. In addition, a study in a European population showed that loss-of-function variants in the ATMgene increased gastric cancer risk [11]. Although the incidence of gastric cancer is high in Korea [12], GWASs targeting the Korean population are limited, and replication phases with a Korean population have been conducted in only a few studies [6,10].
Therefore, we investigated genetic polymorphisms associated with gastric cancer development in a Korean population using a genome-wide scanning approach based on gastric cancer patients and healthy controls and large replication sets from a Korean population cohort.
For GWAS analysis, subjects were recruited from the National Cancer Center (NCC) in Korea. A total of 450 patients with histologically confirmed gastric cancer at the Center for Gastric Cancer between April 2011 and December 2014 and 1,135 controls who participated in a cancer screening program between April 2011 and December 2014 were enrolled in this study. The genomic DNA samples of the participants were extracted from peripheral blood leukocytes.
Genome-wide scanning was performed using Axiom Exome 319 (Affymetrix Inc., Santa Clara, CA) containing 318,983 polymorphisms to 450 gastric cancer cases and 1,135 controls. None of the subjects had a call rate < 90%. Genotyping revealed that 68.1% of the single-nucleotide polymorphisms (SNPs) in the array plate were monomorphic in the Korean population. After excluding monomorphic variants, 86,949 SNPs remained, and SNPs showing (1) call rates < 0.95 in cases or controls (n=0), (2) deviation from the Hardy-Weinberg equilibrium (HWE) (p < 1×10–6) in controls (n=36), or (3) minor allele frequencies < 0.01 (n=36,781) in cases and controls were excluded. There were 40,659 markers remaining, the mean success rate of individuals was 0.998 after filtering, and 450 cases and 1,134 controls were included in the discovery stage.
For replication, 803 gastric cancer cases and 3,693 healthy controls who were recruited as part of the Korean Genome Epidemiology Study (KoGES) from an urban community were used. The genotyping was performed using an Affymetrix Genome-wide Human SNP Array 6.0 (Affymetrix Inc.), and 569,930 variants were available for further analysis after the same quality control criteria were applied.
Genotypes were phased using SHAPEIT (v2. r837), and genotype imputation was performed using IMPUTE2 (2.3.2). The 1000 Genome Project phase 3 East Asian Ancestry sample (n=504) was used for a reference panel. The variants with INFO scores above 0.6 were retained for an NCC sample (n=2,073,558), and for KoGES, variants with INFO ≥ 0.9 (n=6,699,176) were selected. After controlling for the missingness rate, HWE, and minor allele frequency, 713,348 SNPs for NCC and 4,470,730 SNPs for KoGES were available for further analysis.
For the discovery stage, logistic regression analysis based on an additive genetic model was performed to identify the association between each SNP and gastric cancer status with adjustments for age and sex. In addition, we performed stratified analysis by sex. The SNPs with a p-value of < 5×10−5 in the total population or either the male or female subgroup were selected for replication in the KoGES samples. A total of 51 SNPs for the total population, 36 SNPs for males, and 52 SNPs for females were replicated in the KoGES samples. In the replication stage, logistic regression analysis based on an additive genetic model was also applied with adjustment for age and sex. For the replicated SNPs showing statistical significance (p < 0.05 in the replication phase), a codominant model with the homozygote of the major allele as a reference was applied to calculate frequencies and odds ratios (ORs).
In addition, the effect of the PSCA gene was adjusted, and the SNPs with p-value of < 5×10−5 for the total population and males and SNPs with p-value of < 1×10−5 for females in the discovery stage were selected for replication.
To combine the association results for the discovery and replication stages, a meta-analysis with a fixed-effect model was applied. For replicated SNPs showing statistical significance (p < 0.05), regional plots and linkage disequilibrium (LD) are presented. In addition, to compare our results with those of SNPs confirmed to be associated in previous GWASs, ORs of the SNPs that were successfully genotyped or imputed in the GWAS phase in the NCC study population were calculated. These analyses were performed with PLINK v.1.07 [13], R statistical software, METAL [14], and Haplo-View.
A summary of the subjects included in the discovery and replication phase is presented in Table 1. In the GWAS phase, 450 gastric cancer cases including 191 diffuse types, 167 intestinal types, and 92 indeterminate or mixed types, along with 1,134 controls, were included; 803 gastric cancer cases with 394 diffuse types and 409 intestinal types, along with 3,693 controls, were included in replication phase.
In the discovery association analysis adjusted for age and sex, SNPs with p-value of < 5×10−5 were considered for replication. The Manhattan plot and QQ plots of the total, male, and female subjects are presented in S1-S3 Figs. S4-S6 Tables provide the complete results of the discovery and replication phases for the total study subjects, males, and females for the respective 51, 36, and 52 SNPs that showed p-values of < 5×10−5 in the discovery stage, and the corresponding results for these SNPs are also presented for the replication stage and the meta-analysis results. The selected SNPs with p-values of < 5×10−5 in all study subjects and females fell into two large LD blocks. The SNPs in chromosome 8 were located in the PSCA gene, including rs2294008 and 2296392, which were identified in a previous GWAS [6] and were all in the LD block with r2 > 0.9 (Fig. 1).
Table 2 presents up to three SNPs with p-values of < 5×10−5 after LD consideration (if SNPs showed an r2 > 0.7, the most significant SNP was presented). In the case of SNPs located in a large LD block in the PSCA gene, the results of the most significant SNP (rs2976394) and rs2294008, the latter of which is the most commonly reported SNP in the PSCA gene according to previous studies, are presented in the table. In the total population, rs2976394 and rs2294008 in the PSCA gene were associated with gastric cancer with genome-wide significance and showed significant association in the replication phase in the additive model (per increment of C; OR, 0.62; 95% confidence interval [CI], 0.52 to 0.73) in the discovery phase and OR, 0.75; 95% CI, 0.67 to 0.84 in the replication phase). In the NCC population, the rs2976394 TC and CC genotypes were associated with a 0.69- and 0.37-fold decreased cancer risk, respectively, in the codominant model (95% CI, 0.53 to 0.89 and 0.26 to 0.52). In the replication phase, the corresponding ORs were 0.92 (95% CI, 0.77 to 1.10) and 0.53 (95% CI, 0.42 to 0.67). The meta-analysis ORs of the rs2976394 TC and CC genotypes were 0.84 (95% CI, 0.72 to 0.97) and 0.47 (95% CI, 0.39 to 0.57), respectively. The per increment of C in rs2294008 and the TC and CC genotypes also showed decreased gastric cancer risk in the NCC population, the replication phase, and in the meta-analysis results, with association strengths similar to that for rs2976394, reflecting the high LD between these two SNPs.
However, another variant, rs12025565 in chromosome 1, showed an increased risk of 1.45-fold (95% CI, 1.21 to 1.73) per increment of A, 1.72-fold (95% CI, 1.35 to 2.19) for the GA genotype and 1.64-fold (95% CI, 1.06 to 2.55) for the AA genotype in the NCC population but was not replicated in the KoGES population.
In the sex-stratified analysis, rs2976394 and rs2294008 showed similar associations in females, with a significantly replicated association in KoGES females. rs12233126 in chromosome 2 (EIF5B) showed an increased risk (1.77-fold; 95% CI, 1.35 to 2.34, p=4.67E-05) for the AA genotype in the NCC female population but failed to replicate the association. In males, the PSCA gene variants did not show a significant association in the discovery and replication phases, and rs6926644, rs7215433, and rs74396937 were selected but failed to show significance in the replication phase.
We conducted an association analysis after adjusting for known associated SNPs in the PSCA gene (rs2294008 [6]). After considering LD, rs371061408 in chromosome 1 for all subjects; rs6936644, rs7669841, and rs7215433 for male subjects; and rs12233126, rs111673703, and rs10463885 in female subjects were selected with a marginal association (Table 3). However, none of the associations were replicable. The complete results of the discovery and replication phases after adjusting for the PSCA gene for total study subjects, males, and females for 8, 31, and 8 SNPs are shown in S7-S9 Tables.
In Table 4, our discovery stage results are presented for seven SNPs that were investigated in previous gastric cancer GWASs. Five SNPs were not genotyped or imputed in the NCC population. We confirmed a previous GWAS result of an association between SNPs in downstream (rs140081212) or missense variants (rs760077) of the MTX1 gene and the risk of gastric cancer. Another SNP (rs6676150) reported by Helgason et al. [11] was also confirmed. However, we did not observe evidence that other SNPs reported in the GWAS study were also associated in our data.
To the best of the authors’ knowledge, this study is the first GWAS of gastric cancer in a Korean population with a replication population of the same ethnicity [12]. Based on this GWAS analysis, we confirmed that the PSCA gene is a susceptibility gene for gastric cancer in a Korean population.
The PSCA gene encodes a 123-amino acid glycosylphosphatidylinositol-anchored cell membrane glycoprotein related to signal transduction and cell-growth regulation [15,16]. Expression of the PSCA protein is highly expressed and up-regulated in not only prostate but also pancreatic cancers [16,17]. However, PSCA expression was reduced in the bladder, esophagus, and gastric cancer [6,18], and the downregulation was more prominent in diffuse types of gastric cancer [6]. The association between the PSCA gene and gastric cancer susceptibility was first identified in a GWAS in a Japanese population with a more prominent effect in diffuse-type gastric cancer, and these results were also well replicated in a Korean population. In functional assays, PSCA inhibited cell proliferation and induced cell death, and the rs2294008 C>T transition reduced the transcriptional activity of PSCA; however, other risk alleles in PSCA did not change transcriptional activity, suggesting that there was a modulator effect for the rs2294008 SNP on PSCA promoter activity [6]. Following the GWAS, the association of rs2294008 with gastric cancer has been investigated extensively, and recent meta-analyses showed that the T allele of rs2294008 in the PSCA gene increased gastric cancer risk, especially for non-cardiac or diffuse gastric cancer, in an Asian population [19,20]. A recent study in a Caucasian population showed similar results in gastric cancer development and determined that rs2294008C>T was associated with worse prognosis in diffuse-type gastric cancer patients [21].
In this study, we confirmed the association of the PSCA gene and gastric cancer risk with genome-wide significance. When the minor allele (C) was considered as a risk allele, it decreased gastric cancer risk, while the T allele increased the gastric cancer risk. The most significant SNP in this study was rs2976394, with strong LD in rs2294008 (r2 > 0.95). A previous candidate SNP approach in Korean gastric cancer cases and controls also showed an increased association of the T allele of rs2294008 with gastric cancer risk [22]. The selected and replicated SNPs with statistical significance in this study were all included in the LD block of the PSCA gene. However, when we stratified the analysis according to sex, the association of SNPs in the PSCA gene, including rs2294008 and 2976394, was more prominent in females. In males, rs2976394 showed a p-value of 3×10−4, and rs2294008 had a p-value of 6×10−4 (data not shown). However, the association of rs2294008 between males and females did not differ in a previous Korean study [22]. In Korea, the behavioral risk factors for gastric cancer differ by sex. Although the H. pylori infection rates were similar between males and females [23], the smoking rate was much higher in males (42.1% vs. 6.2%), and sodium and processed meat intake were higher in the male population [24]. These differences in behavioral patterns between males and females might affect the significance of the PSCA gene on gastric cancer. Further studies to identify sex differences may be needed. When gastric cancer was divided by histological type, i.e., diffuse and intestinal type, the association of polymorphisms in the PSCA gene was more prominent in the diffuse type. For rs2294008, the ORs per increment of the C allele were 0.50 (95% CI, 0.39 to 0.63; p=1.98×10−8) in the discovery phase and 0.66 (95% CI, 0.57 to 0.77; p=1.43×10−7) in the replication phase for diffuse type cancer, similar to the results of a previous GWAS [6] and other studies [19,20]. Otherwise, in intestinal type cancer, the ORs per increment of the C allele were 1.23 (95% CI, 0.97 to 1.57; p=0.08) in the discovery phase result and 0.85 (95% CI, 0.73 to 0.99, p=0.03) in the replication phase.
Compared to previous GWASs, this study showed a significant association between variants in the MTX1 gene and gastric cancer (rs760077 and rs140081212), as also reported by Helgason et al. [11]. The MTX gene encodes a mitochondrial protein [25], and in terms of gastric cancer risk-related functions, the MTX1 gene is classified as an apoptosis and proliferation gene [26]. The MTX1 gene showed strong LD with the MUC1 gene [11,26]. Helgason et al. [11] also found that rs760077 was perfectly correlated with rs4072037, which has an effect on alternative splicing of MUC1. Although we did not find an association for rs4072037 (MUC1) in our study subjects (Table 4), we identified the association between rs2070803, another polymorphism downstream of both MUC1 and the ITRIM46 gene [26], and gastric cancer. ITRIM-46 also showed association with gastric cancer (p=0.02, data not shown), suggesting plausible associations of rs760077 and rs140081212 polymorphisms in this population. Among the SNPs studied in prior GWASs, a previous candidate SNP approach in a Korean population demonstrated significant associations of rs10074991, 13361707 (PRKAA1) [27], rs407-2037 (MUC1), and rs2274223 (PLCE) [28], but we did not find associations of these polymorphisms with gastric cancer. In addition, another replication study of six SNPs from previous GWASs also identified that only two genes—PRKAA1 and PSCA—showed significant associations in the Korean population [29].
Several limitations of this study need to be mentioned. First, in this study, information about environmental factors, such as smoking, H. pylori infection, diet, and family history, were not considered, although these are significant risk factors for gastric cancer [3,4]. However, considering the definition of confounder, variations in genes as explanatory variables are unaffected by environmental factors. Therefore, the environmental factors that were not considered may not be confounders, although they could act as intermediate variables between SNPs and gastric cancer [6]. Second, the number of subjects in this study was relatively smaller than the number in previous GWASs, and the statistical power was limited. Despite these limitations, this is the first GWAS with genetically homogeneous replication subjects from a Korean population. In addition, conditional analysis of the PSCA gene may confirm that PSCA is the most important genetic variation associated with gastric cancer in a Korean population.
In summary, we confirmed that variants in the PSCA gene showed significant GWAS associations in a Korean population for gastric cancer, especially for females and diffuse type gastric cancer. In addition, polymorphisms in the MTX1 gene showed associations with gastric cancer, possibly due to correlation with the MUC1 gene.
Electronic Supplementary Material
Supplementary materials are available at Cancer Research and Treatment website (http://www.e-crt.org).
ACKNOWLEDGMENTS
This work was supported by grants from the National Cancer Center, Republic of Korea (no. 1410260, 1810980).
This study was provided with biospecimens and data from the Korean Genome Analysis Project (4845-301), the Korean Genome and Epidemiology Study (4851-302), and Korea Biobank Project (4851-307, KBP-2015-053) that were supported by the Korea Center for Disease Control and Prevention, Republic of Korea.
References
1. Global Burden of Disease Cancer Collaboration, Fitzmaurice C, Dicker D, Pain A, Hamavid H, Moradi-Lakeh M, et al. The Global Burden of Cancer 2013. JAMA Oncol. 2015; 1:505–27.
2. Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010; 127:2893–917.

3. Compare D, Rocco A, Nardone G. Risk factors in gastric cancer. Eur Rev Med Pharmacol Sci. 2010; 14:302–8.
4. Palli D. Epidemiology of gastric cancer: an evaluation of available evidence. J Gastroenterol. 2000; 35 Suppl 12:84–9.
5. Singh K, Ghoshal UC. Causal role of Helicobacter pylori infection in gastric cancer: an Asian enigma. World J Gastroenterol. 2006; 12:1346–51.

6. Study Group of Millennium Genome Project for Cancer, Sakamoto H, Yoshimura K, Saeki N, Katai H, Shimoda T, et al. Genetic variation in PSCA is associated with susceptibility to diffuse-type gastric cancer. Nat Genet. 2008; 40:730–40.

7. Abnet CC, Freedman ND, Hu N, Wang Z, Yu K, Shu XO, et al. A shared susceptibility locus in PLCE1 at 10q23 for gastric adenocarcinoma and esophageal squamous cell carcinoma. Nat Genet. 2010; 42:764–7.

8. Shi Y, Hu Z, Wu C, Dai J, Li H, Dong J, et al. A genome-wide association study identifies new susceptibility loci for non-cardia gastric cancer at 3q13.31 and 5p13.1. Nat Genet. 2011; 43:1215–8.

9. Jin G, Ma H, Wu C, Dai J, Zhang R, Shi Y, et al. Genetic variants at 6p21.1 and 7p15.3 are associated with risk of multiple cancers in Han Chinese. Am J Hum Genet. 2012; 91:928–34.

10. Hu N, Wang Z, Song X, Wei L, Kim BS, Freedman ND, et al. Genome-wide association study of gastric adenocarcinoma in Asia: a comparison of associations between cardia and noncardia tumours. Gut. 2016; 65:1611–8.

11. Helgason H, Rafnar T, Olafsdottir HS, Jonasson JG, Sigurdsson A, Stacey SN, et al. Loss-of-function variants in ATM confer risk of gastric cancer. Nat Genet. 2015; 47:906–10.

12. Leung WK, Wu MS, Kakugawa Y, Kim JJ, Yeoh KG, Goh KL, et al. Screening for gastric cancer in Asia: current evidence and practice. Lancet Oncol. 2008; 9:279–87.

13. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007; 81:559–75.

14. Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient metaanalysis of genomewide association scans. Bioinformatics. 2010; 26:2190–1.

15. Sharom FJ, Radeva G. GPI-anchored protein cleavage in the regulation of transmembrane signals. Subcell Biochem. 2004; 37:285–315.

16. Reiter RE, Gu Z, Watabe T, Thomas G, Szigeti K, Davis E, et al. Prostate stem cell antigen: a cell surface marker overexpressed in prostate cancer. Proc Natl Acad Sci U S A. 1998; 95:1735–40.

17. Grubbs EG, Abdel-Wahab Z, Tyler DS, Pruitt SK. Utilizing quantitative polymerase chain reaction to evaluate prostate stem cell antigen as a tumor marker in pancreatic cancer. Ann Surg Oncol. 2006; 13:1645–54.

18. Bahrenberg G, Brauers A, Joost HG, Jakse G. Reduced expression of PSCA, a member of the LY-6 family of cell surface antigens, in bladder, esophagus, and stomach tumors. Biochem Biophys Res Commun. 2000; 275:783–8.
19. Shi D, Wang S, Gu D, Wu D, Wang M, Chu H, et al. The PSCA polymorphisms derived from genome-wide association study are associated with risk of gastric cancer: a meta-analysis. J Cancer Res Clin Oncol. 2012; 138:1339–45.

20. Wang T, Zhang L, Li H, Wang B, Chen K. Prostate stem cell antigen polymorphisms and susceptibility to gastric cancer: a systematic review and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2012; 21:843–50.

21. Garcia-Gonzalez MA, Bujanda L, Quintero E, Santolaria S, Benito R, Strunk M, et al. Association of PSCA rs2294008 gene variants with poor prognosis and increased susceptibility to gastric cancer and decreased risk of duodenal ulcer disease. Int J Cancer. 2015; 137:1362–73.
22. Song HR, Kim HN, Piao JM, Kweon SS, Choi JS, Bae WK, et al. Association of a common genetic variant in prostate stemcell antigen with gastric cancer susceptibility in a Korean population. Mol Carcinog. 2011; 50:871–5.

23. Lim SH, Kwon JW, Kim N, Kim GH, Kang JM, Park MJ, et al. Prevalence and risk factors of Helicobacter pylori infection in Korea: nationwide multicenter study over 13 years. BMC Gastroenterol. 2013; 13:104.

24. Wang M, Zhang R, He J, Qiu L, Li J, Wang Y, et al. Potentially functional variants of PLCE1 identified by GWASs contribute to gastric adenocarcinoma susceptibility in an eastern Chinese population. PLoS One. 2012; 7:e31932.

25. Gan-Or Z, Bar-Shira A, Gurevich T, Giladi N, Orr-Urtreger A. Homozygosity for the MTX1 c.184T>A (p.S63T) alteration modifies the age of onset in GBA-associated Parkinson's disease. Neurogenetics. 2011; 12:325–32.
26. Mocellin S, Verdi D, Pooley KA, Nitti D. Genetic variation and gastric cancer risk: a field synopsis and meta-analysis. Gut. 2015; 64:1209–19.

27. Kim YD, Yim DH, Eom SY, Moon SI, Yun HY, Song YJ, et al. Risk of gastric cancer is associated with PRKAA1 gene polymorphisms in Koreans. World J Gastroenterol. 2014; 20:8592–8.
Fig. 1.
Linkage disequilibrium plot and regional plot of rs2294008 in study subjects in the discovery phase. (A) Linkage disequilibrium plot. (B) Regional plot.
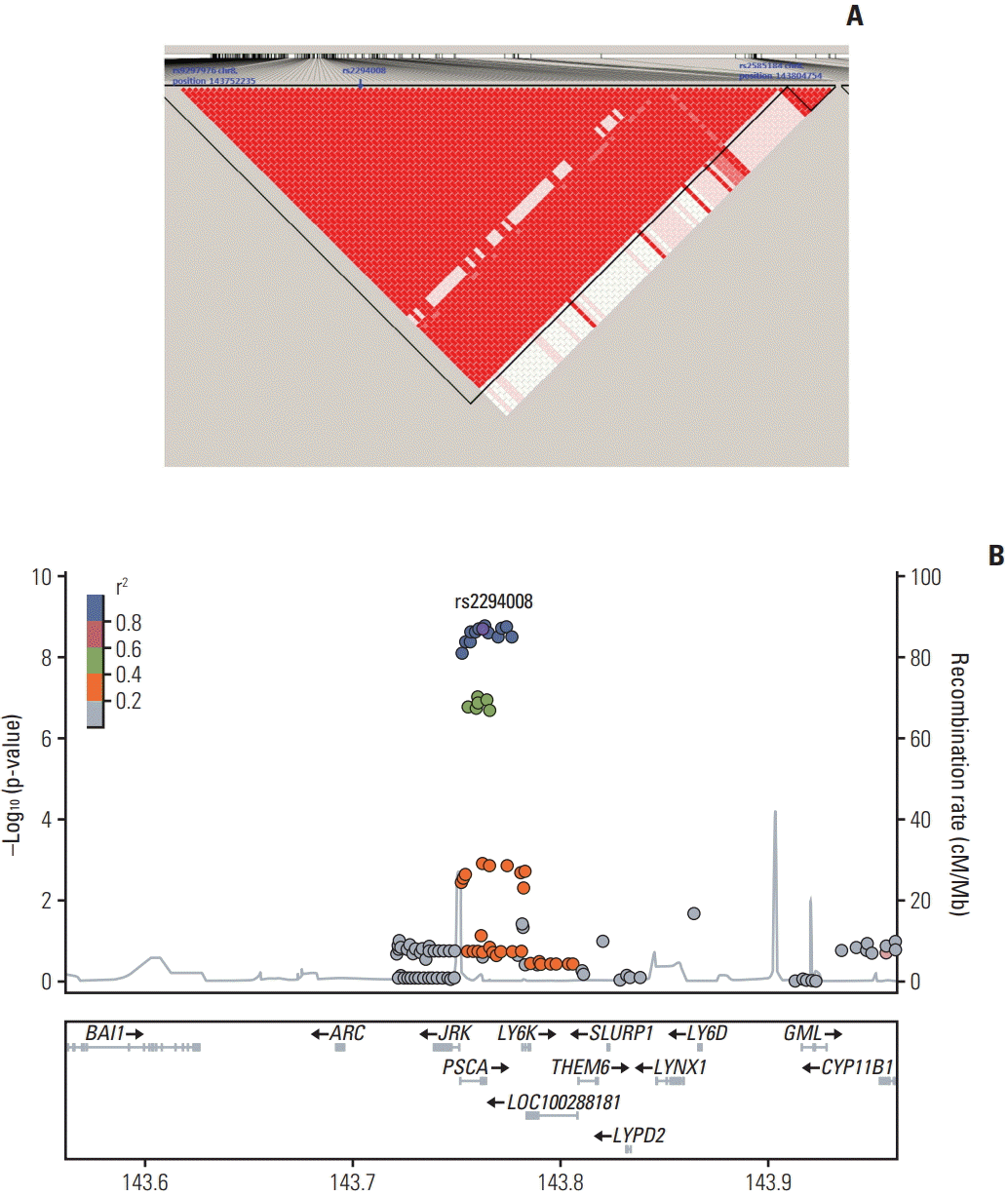
Table 1.
Summary demographic description of the study subjects
Table 2.
SNPs associated with gastric cancer in both the genome-wide association stage and replication study
| SNP | Chr | Gene | Genotype frequency |
GWAS stage |
Replication |
Meta-analysis |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case Afreq | Cont Afreq | p-value | OR (95% CI) | Case Afreq | Cont Afreq | p-value | OR (95% CI) | p-value | OR (95% CI) | |||||
| Total population | ||||||||||||||
| rs2976394a) | 8 | PSCA | Tb)Cc) | 0.40 | 0.52 | 1.07E-08 | 0.62 (0.52-0.73) | 0.43 | 0.50 | 5.09E-07 | 0.75 (0.67-0.84) | 1.66E-13 | 0.70 (0.64-0.77) | |
| TT | 0.34 | 0.23 | Ref | 1 | 0.29 | 0.25 | Ref | 1 | Ref | 1 | ||||
| TC | 0.51 | 0.50 | 0.00427 | 0.69 (0.53-0.89) | 0.55 | 0.50 | 0.351 | 0.92 (0.77-1.10) | 0.0155 | 0.84 (0.72-0.97) | ||||
| CC | 0.14 | 0.27 | 9.86E-09 | 0.37 (0.26-0.52) | 0.16 | 0.25 | 1.61E-07 | 0.53 (0.42-0.67) | < 0.0001 | 0.47 (0.39-0.57) | ||||
| rs2294008d) | 8 | PSCA | Tb)Cc) | 0.40 | 0.52 | 1.38E-08 | 0.62 (0.53-0.73) | 0.43 | 0.50 | 5.28E-07 | 0.75 (0.67-0.84) | 1.94E-13 | 0.70 (0.64-0.77) | |
| TT | 0.35 | 0.23 | Ref | 1 | 0.29 | 0.25 | Ref | 1 | Ref | 1 | ||||
| TC | 0.51 | 0.50 | 0.051 | 0.69 (0.54-0.90) | 0.55 | 0.50 | 0.348 | 0.92 (0.76-1.10) | 0.0164 | 0.85 (0.72-0.97) | ||||
| CC | 0.14 | 0.26 | 1.23E-08 | 0.37 (0.26-0.52) | 0.16 | 0.25 | 1.67E-07 | 0.53 (0.41-0.67) | < 0.0001 | 0.47 (0.39-0.57) | ||||
| rs12025565e) | 1 | - | Gb)Ac) | 0.32 | 0.24 | 4.47E-05 | 1.45 (1.21-1.73) | 0.28 | 0.27 | 0.3841 | 1.06 (0.93-1.20) | 0.0023 | 1.17 (1.06-1.30) | |
| GG | 0.44 | 0.57 | Ref | 1 | 0.51 | 0.54 | Ref | 1 | Ref | 1 | ||||
| GA | 0.45 | 0.36 | 9.39E-06 | 1.72 (1.35-2.19) | 0.42 | 0.39 | 0.087 | 1.15 (0.97-1.36) | < 0.001 | 1.31 (1.15-1.50) | ||||
| AA | 0.09 | 0.07 | 0.0261 | 1.64 (1.06-2.55) | 0.07 | 0.07 | 0.808 | 0.96 (0.70-1.32) | 0.2115 | 1.18 (0.91-1.61) | ||||
| Male | ||||||||||||||
| rs6936644f) | 6 | ZSCAN31 | Cb)Tc) | 0.54 | 0.42 | 9.60E-06 | 1.57 (1.29-1.92) | 0.49 | 0.47 | 0.3367 | 1.07 (0.93-1.24) | 0.0008 | 1.22 (1.09-1.37) | |
| CC | 0.22 | 0.35 | Ref | 1 | 0.24 | 0.26 | Ref | 1 | Ref | 1 | ||||
| CT | 0.48 | 0.46 | 0.002785 | 1.71 (1.20-2.44) | 0.53 | 0.52 | 0.328 | 1.13 (0.89-1.43) | 0.0155 | 1.27 (1.05-1.55) | ||||
| TT | 0.30 | 0.19 | 1.42E-05 | 2.45 (1.63-3.67) | 0.22 | 0.22 | 0.357 | 1.14 (0.86-1.52) | 0.0011 | 1.47 (1.17-1.86) | ||||
| rs7215433g) | 17 | - | Tb)Cc) | 0.20 | 0.29 | 2.80E-05 | 0.59 (0.46-0.75) | 0.23 | 0.24 | 0.6649 | 0.96 (0.82-1.14) | 0.0081 | 0.83 (0.73-0.95) | |
| TT | 0.64 | 0.49 | Ref | 1 | 0.60 | 0.58 | Ref | 1 | Ref | 1 | ||||
| TC | 0.33 | 0.43 | 0.000343 | 0.57 (0.42-0.78) | 0.34 | 0.36 | 0.436 | 0.92 (0.75-1.14) | 0.0069 | 0.79 (0.66-0.94) | ||||
| CC | 0.04 | 0.07 | 0.007788 | 0.39 (0.19-0.78) | 0.06 | 0.06 | 0.908 | 1.02 (0.68-1.55) | 0.1962 | 0.79 (0.55-1.13) | ||||
| rs74396937 | 11 | OR4C46 | Ab)Gc) | 0.08 | 0.14 | 3.65E-05 | 0.49 (0.35-0.69) | 0.25 | 0.26 | 0.3150 | 0.92 (0.79-1.08) | 0.0079 | 0.82 (0.71-0.95) | |
| AA | 0.86 | 0.74 | Ref | 1 | 0.58 | 0.54 | Ref | 1 | Ref | 1 | ||||
| AG | 0.12 | 0.24 | 5.72E-06 | 0.39 (0.26-0.58) | 0.35 | 0.39 | 0.159 | 0.86 (0.70-1.06) | 0.0016 | 0.74 (0.62-0.89) | ||||
| GG | 0.02 | 0.03 | 0.235 | 0.55 (0.21-1.47) | 0.07 | 0.07 | 0.831 | 0.96 (0.65-1.42) | 0.6295 | 0.92 (0.64-1.31) | ||||
| Female | ||||||||||||||
| rs2976394a) | 8 | PSCA | Tb)Cc) | 0.38 | 0.53 | 3.36E-06 | 0.53 (0.40-0.69) | 0.43 | 0.50 | 0.0043 | 0.77 (0.64-0.92) | 6.71E-07 | 0.68 (0.58-0.79) | |
| TT | 0.35 | 0.22 | Ref | 1 | 0.29 | 0.25 | Ref | 1 | Ref | 1 | ||||
| TC | 0.54 | 0.50 | 0.036255 | 0.65 (0.43-0.97) | 0.55 | 0.50 | 0.799 | 0.96 (0.72-1.29) | 0.1761 | 0.85 (0.67-1.08) | ||||
| CC | 0.11 | 0.28 | 5.55E-06 | 0.25 (0.14-0.46) | 0.16 | 0.25 | 0.002 | 0.54 (0.37-0.81) | < 0.0001 | 0.42 (0.31-0.59) | ||||
| rs2294008d) | 8 | PSCA | Tb)Cc) | 0.38 | 0.53 | 5.10E-06 | 0.53 (0.41-0.70) | 0.43 | 0.50 | 0.0043 | 0.76 (0.64-0.92) | 8.00E-07 | 0.68 (0.59-0.79) | |
| TT | 0.35 | 0.22 | Ref | 1 | 0.29 | 0.25 | Ref | 1 | Ref | 1 | ||||
| TC | 0.43 | 0.50 | 0.05599 | 0.67 (0.45-1.01) | 0.55 | 0.50 | 0.838 | 0.97 (0.72-1.30) | 0.2281 | 0.86 (0.68-1.10) | ||||
| CC | 0.11 | 0.28 | 7.39E-06 | 0.26 (0.14-0.46) | 0.15 | 0.25 | 0.002 | 0.54 (0.36-0.80) | < 0.0001 | 0.42 (0.30-0.59) | ||||
| rs12233126f) | 2 | EIF5B | Tb)Ac) | 0.38 | 0.27 | 4.97E-05 | 1.77 (1.34-2.33) | 0.28 | 0.28 | 0.9352 | 1.01 (0.82-1.24) | 0.0135 | 1.23 (1.04-1.45) | |
| TT | 0.38 | 0.54 | Ref | 1 | 0.52 | 0.51 | Ref | 1 | Ref | 1 | ||||
| TA | 0.48 | 0.39 | 0.003291 | 1.80 (1.21-2.66) | 0.40 | 0.42 | 0.851 | 0.97 (0.75-1.27) | 0.1753 | 1.16 (0.93-1.45) | ||||
| AA | 0.14 | 0.07 | 0.000249 | 3.11 (1.69-5.69) | 0.08 | 0.07 | 0.769 | 1.08 (0.66-1.76) | 0.0185 | 1.57 (1.07-2.30) | ||||
Table 3.
SNPs associated with gastric cancer in both the genome-wide association stage and replication study after conditioning on the PSCA gene (rs2294008)
| SNP | Chr | Gene | Maj | Min |
GWAS stage |
Replication |
Meta-analysis |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case Afreq | Cont Afreq | p-value | OR (95% CI) | Case Afreq | Cont Afreq | p-value | OR (95% CI) | p-value | OR (95% CI) | ||||||
| Total population | |||||||||||||||
| rs371061408a) | 1 | - | T | C | 0.32 | 0.24 | 3.72E-05 | 1.46 (1.22-1.75) | 0.28 | 0.27 | 0.3395 | 1.06 (0.94-1.21) | 0.0017 | 1.18 (1.06-1.31) | |
| Male | |||||||||||||||
| rs6936644b) | 6 | ZSCAN31 | C | T | 0.49 | 0.48 | 1.81E-05 | 1.56 (1.27-1.90) | 0.44 | 0.48 | 0.3231 | 1.07 (0.93-1.24) | 0.0010 | 1.22 (1.08-1.37) | |
| rs7669841b) | 4 | TET2 | G | C | 0.30 | 0.30 | 3.64E-05 | 1.58 (1.27-1.97) | 0.31 | 0.30 | 0.2850 | 0.92 (0.79-1.07) | 0.1405 | 1.10 (0.97-1.24) | |
| rs7215433c) | 17 | - | T | C | 0.26 | 0.28 | 3.80E-05 | 0.59 (0.46-0.76) | 0.25 | 0.26 | 0.5488 | 0.95 (0.81-1.12) | 0.0060 | 0.83 (0.72-0.95) | |
| Female | |||||||||||||||
| rs12233126d) | 2 | EIF5B | T | A | 0.38 | 0.27 | 6.27E-05 | 1.77 (1.34-2.35) | 0.28 | 0.28 | 0.8231 | 1.02 (0.83-1.26) | 0.0109 | 1.24 (1.05-1.46) | |
| rs111673703b) | 11 | APLP2 | C | T | 0.11 | 0.05 | 7.08E-05 | 2.49 (1.59-3.91) | 0.07 | 0.06 | 0.6472 | 1.09 (0.75-1.59) | 0.0037 | 1.53 (1.15-2.05) | |
| rs10463885 | 5 | CDC42SE2 | A | G | 0.06 | 0.02 | 9.73E-05 | 4.17 (2.03-8.55) | 0.02 | 0.03 | 0.6665 | 0.88 (0.48-1.6) | 0.0289 | 1.68 (1.05-2.66) | |
Table 4.
Associations from the genome-wide association stage data for SNPs showing significance in previously reported GWAS studies
| SNP | Chr | Gene |
GWAS in previous report |
GWAS stage in this study |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ethnicity | Effect | Cont Afreq | p-value | OR (95% CI) | Maj | Min | Case Afreq | Cont Afreq | p-value | OR (95% CI) | |||
| rs140081212 [11] | 1 | - | European (Icelanders) | A | 0.65 | 7.90E-10 | 0.79 (0.73-0.86) | G | A | 0.09 | 0.12 | 0.0366 | 0.73 (0.55-0.96) |
| rs760077 [11] | 1 | MTX1 | European (Icelanders) | A | 0.65 | 1.10E-08 | 0.79 (0.73-0.85) | T | A | 0.09 | 0.11 | 0.0152 | 0.71 (0.54-0.94) |
| rs6676150 [11] | 1 | - | European (Icelanders) | C | 0.67 | 1.50E-08 | 0.80 (0.74-0.86) | G | C | 0.06 | 0.10 | 0.0018 | 0.61 (0.44-0.83) |
| rs2274223[7] | 10 | PLCE | Asian (Chinese) | G | 0.21 | 8.40E-09 | 1.31 (1.19-1.43) | C | T | 0.26 | 0.26 | 0.4074 | 1.08 (0.90-1.30) |
| rs3765524[7] | 10 | PLCE | Asian (Chinese) | T | 0.21 | 5.32E-09 | 1.31 (1.20-1.44) | C | T | 0.26 | 0.26 | 0.5340 | 1.08 (0.90-1.30) |
| rs9841504[8] | 3 | Asian (Han Chinese) | G | 0.15 | 1.70E-09 | 0.76 (0.69-0.83) | - | - | - | - | - | - | |
| rs13361707 [8] | 5 | Asian (Han Chinese) | C | 0.48 | 7.60E-29 | 1.41 (1.32-1.49) | - | - | - | - | - | - | |
| rs10074991 [10] | 5 | PRKAA1 | Asian (China, Korea) | A | 0.48 | 4.83E-26 | 0.80 (0.77-0.83) | - | - | - | - | - | - |
| rs2294693 [10] | 6 | UNC5CL, TSPO2 | Asian (China, Korea) | C | 0.24 | 7.22E-08 | 1.14 (1.09-1.20) | - | - | - | - | - | - |
| rs4072037 [10] | 1 | MUC1 | Asian (China, Korea) | G | NR | 7.00E-08 | 1.32 (1.19-1.45) | T | C | 0.12 | 0.14 | 0.2070 | 0.82 (0.64-1.05) |
| rs2494938[9] | 6 | LRFN2 | Asian (Han Chinese) | A | 0.26 | 4.91E-09 | 1.18 (1.12-1.25) | - | - | - | - | - | - |
| rs2285947[9] | 7 | DNAH11 | Asian (Han Chinese) | A | 0.27 | 1.36E-06 | 1.14 (1.08-1.21) | G | A | 0.32 | 0.32 | 0.7298 | 0.99 (0.84-1.17) |




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download