Abstract
Purpose
In the presence of interferon, proteasome subunits are replaced by their inducible counterparts to form an immunoproteasome (IP) plays a key role in generation of antigenic peptides presented by MHC class I molecules, leading to elicitation of a T cell‒mediated immune response. Although the roles of IP in other cancers, and inflammatory diseases have been extensively studied, its significance in breast cancer is unclear.
Materials and Methods
We investigated the expression of LMP7, an IP subunit, and its relationship with immune system components in two breast cancer cohorts.
Results
In 668 consecutive breast cancer cohort, 40% of tumors showed high level of LMP7 expression, and tumors with high expression of LMP7 had more tumor-infiltrating lymphocytes (TILs) in each subtype of breast cancer. In another cohort of 681 triple-negative breast cancer patients cohort, the expression of LMP7 in tumor cells was significantly correlated with the amount of TILs and the expression of interferon-associated molecules (MxA [p < 0.001] and PKR [p < 0.001]), endoplasmic reticulum stress-associated molecules (PERK [p=0.012], p-eIF2a [p=0.001], and XBP1 [p < 0.001]), and damage-associated molecular patterns (HMGN1 [p < 0.001] and HMGB1 [p < 0.001]). Patients with higher LMP7 expression had better disease-free survival outcomes than those with no or low expression in the positive lymph node metastasis group (p=0.041).
Triple-negative breast cancers (TNBCs) have a poor prognosis and low survival rate than other subtypes. Until now, no standard targeted treatment for TNBCs is available. Currently, much attention is focused on immunotherapy-related treatments [1]. The effects of conventional breast cancer treatments are partly due to interactions with the immune system. Therefore, improved insight into the connection between the immune system and breast cancer may improve treatments and outcomes.
Proteasomes are multisubunit complexes that degrade intracellular proteins through the ubiquitin–proteasome pathway [2,3]. In the presence of interferon, the three catalytic β subunits β1, β2, and β5 in the proteasome complex are replaced by proteasome subunit LMP2 (PSMB9)/β1i, LMP10 (PSMB10)/β2i, and LMP7 (PSMB8)/β5i, respectively, to form the immunoproteasome (IP). Subunits β5 and LMP7 perform chymotrypsin-like activity, subunits β1 and LMP2 perform caspase-like activity, and subunits β2 and LMP10 perform trypsin-like activity. However, the substrate-binding pockets of IPs differ from those of proteasome. IPs display an altered cleavage patterns, generating peptides suitable for binding onto a major histocompatibility complex (MHC) class I, facilitating CD8+ T-cell responses [4]. Thus, IPs play a key role for T cell–mediated immunity by contributing antigen epitopes presentation in tumors. Lack of expression or downregulation of an IP may contribute to immune evasion through antigen loss.
We previously demonstrated that elevated endoplasmic reticulum stress in TNBCs triggers release of damage-associated molecular patterns such as HMGB1 and HMGN1, which is known to be associated with attraction of tumor-infiltrating lymphocytes (TILs) to tumor microenvironment [5-9]. Then TILs can release interferon, which can elevate expression of MHC classes on tumor cells. However, the impact of IP expression on antigen presentation in breast cancer is not well established. In this present study, we evaluated the expression of LMP7 in a consecutive breast cancer and another TNBC tissue series. The higher the LMP7 level was, the higher the TILs were in each subtype of breast cancer. Also in TNBC cohort, higher levels of LMP7 were correlated with higher histologic grades and greater TILs. We also found that LMP7 was a better prognostic factor for disease-free survival in the group of positive lymph node metastases of the TNBC cohort.
Two sets of breast cancer samples were used in the current analyses. The first series included 668 consecutive breast cancer patients who underwent surgery for primary breast cancer in 1998 at Asan Medical Center, Seoul, Korea and for whom formalin-fixed, paraffin-embedded tissue samples were available. Six cycles of an adjuvant cyclophosphamide (500 mg/m2), methotrexate (40 mg/m2), and 5-fluorouracil (500 mg/m2) or cyclophosphamide (500 mg/m2), adriamycin (50 mg/m2), and 5-fluorouracil (500 mg/m2) were administered in 439 of the 668 patients in the consecutive breast cancer cohort. Twelve patients underwent adjuvant chemotherapy with unknown regimens. Radiotherapy and hormone therapy was performed in 178 (26.6%) and 439 (65.7%) patients, respectively. The median follow-up duration was 13.3 years.
We evaluated 681 TNBC patients with surgically resected tissues obtained from surgeries conducted between 2004 and 2010 at Asan Medical Center, and 16 were excluded due to a lack of a tissue sample for tissue microarray analyses. Thus, 665 breast cancer patients were available for immunohistochemistry for LMP7. In this group, 476 patients did not present with lymph node metastasis, and they received four cycles of adjuvant adriamycin (60 mg/m2) plus cyclophosphamide (600 mg/m2) (AC). The remaining 205 patients presented with lymph node metastases and were treated with four cycles of AC followed by either four cycles of docetaxel (75 mg/m2) or paclitaxel (175 mg/m2). In total, 537 patients (80.8%) received radiotherapy. The median follow-up period was 78.3 months. Clinical information was obtained from medical records and pathology reports from the surgery.
The hematoxylin and eosin-stained slides were reviewed by two pathologists (H.J.L. and G.G.). The histologic type of breast cancer was assessed based on the 2012 World Health Organization classification, and the histologic grade of breast cancer was evaluated according to the modified Bloom-Richardson grading system. The slides were evaluated for the levels of stromal TILs using full sections in 10% increments (if less than 10%, 1%, or 5% level criteria were used) as well as the histologic subtype and grade, pT stage, and pN stage.
Formalin-fixed, paraffin-embedded tissue samples were arrayed with a tissue-arraying instrument as previously described [10]. Tissue microarray sections were stained with an automatic immunohistochemical staining device (Benchmark XT, Ventana Medical Systems, Tucson, AZ). Antibodies for LMP7 (1:2,000, ab95926, Santa Cruz Biotechnology, Santa Cruz, CA), human leukocyte antigen (HLA)-ABC (1:1,600, ab70328, EMR8–5, Abcam, Cambridge, UK), estrogen receptor (ER; 1:200, Leica Biosystems, Newcastle upon Tyne, UK), progesterone receptor (PR; 1:200, Leica Biosystems), human epidermal growth factor receptor 2 (HER2; 1:8, Ventana Medical Systems), MxA (1:1,000, ab95926, Abcam), PERK (1:200, Cell Signaling Technology, Danvers, MA), p-eIF2α (1:200, ab32157, Abcam), XBP1 (1:75, ab37152, Abcam), PKR (1:400, ab32052, Abcam), HMGB1 (1:200, ab18256, Abcam), and HMGN1 (1:1,000, ab5212, Abcam) were used. ER and PR levels were considered positive if at least 1% positive tumor nuclei staining is identified [11]. Hormone receptor-positive (HR+) tumors were classified if those were positive on ER and/or PR staining. HER2-overexpressing tumors were classified if those were 3+ scores by immunohistochemistry or positive gene amplification by silver in situ hybridization [12]. The intensity of immunochemical staining was evaluated on a four-value intensity scale (0, 1, 2, and 3). The percentages of membranous and/or cytoplasmic expression were also evaluated. The immunoreactive score was calculated as multiply the product of the intensity by the percentage of positive cells.
HLA-ABC expression in tumor cells were categorized as one of three levels, as previously described (strongly positive, expression in more than 75% of tumor cells; weakly positive, expression between 25% and 75% of tumor cells; negative, loss of expression in more than 75% of tumor cells) [13].
The Cancer Genome Atlas (TCGA) data portal was used to download breast cancer data [14]. A total of 84 cases of basal-like breast cancer were analyzed. We downloaded level-3 gene expression data derived from an Agilent custom 244K whole genome microarray and somatic mutation data from an Illumina Genomic Analyzer (San Diego, CA). The gene expression level was median-centered by gene.
All statistical analyses were carried out using SPSS ver. 18 (SPSS Inc., Chicago, IL) and R statistical software. The Kruskal-Wallis test, chi-squared test, Spearman’s correlation, log-rank test, Cox proportional hazards regression model, and hierarchical clustering were used, as appropriate. All tests were two-sided and were done at significance level of α=0.05.
To determine the histopathologic characteristics of the breast cancers according to LMP7 expression, we analyzed 668 consecutive breast tumor tissues of the first consecutive breast cancer series consisting of 373 HR+/HER2–, 44 HR+/HER2+, 81 HR–/HER2+, and 190 TNBC tumors (Table 1). A loss of LMP7 expression in tumor cells was present in 181 cases, while 218 and 269 tumors showed low and high levels of LMP7 expression, respectively (Fig. 1). LMP7 expression level was similar in each breast cancer subtype (immunoreactive score: HR+/HER2–, 54.1±64.7; HR+/HER2+, 46.5±57.9; HR–/HER2+, 50.1±63.5; TNBC, 62.0±67.5). Tumors with high levels of LMP7 expression had more TILs (p ≤ 0.041) in all subtypes. Since LMP7 expression was correlated with the level of TILs, we further analyzed the association of LMP7 expression with other factors that were correlated with the level of TILs in previous studies [5-9]. LMP7 expression in tumor cells was significantly correlated with interferon-associated molecules HLA-ABC (p ≤ 0.007) and MxA (p < 0.001, except HR+/HER2+ subtype). Higher expression of endoplasmic reticulum stress-associated molecule p-eIF2a was significantly associated with high level of LMP7 in HR+/HER2– subtype (p=0.008). Cytoplasmic expression of damage associated molecular pattern HMGN1 was higher in tumors with high LMP7 (p=0.01). However, the histologic grade, pT stage, and lymph node metastases were not significantly associated with LMP7 expression. In survival analysis, LMP7 expression was not associated with disease-free survival in all subtypes (HR+/HER2–: p=0.61; hazard ratio, 0.999; 95% confidence interval [CI], 0.996 to 1.002; HR+/HER2+: p=0.143; hazard ratio, 1.006, 95% CI, 0.998 to 1.013; HR–/HER2+: p=0.307; hazard ratio, 1.003; 95% CI, 0.998 to 1.008; TNBC: p=0.158; hazard ratio, 0.996; 95% CI, 0.992 to 1.001).
Since TNBCs are aggressive and considered as appropriate candidates for cancer immunotherapy among breast cancer subtypes, we further analyzed LMP7 expression in another cohort of 681 TNBCs, which had homogeneous standardized adjuvant treatment. The expression of LMP7 was analyzable in 665 cases: 20.6%, 38.6%, and 40.8% of TNBCs showed no, low, and high cytoplasmic expression of LMP7, respectively (Table 2). In TNBCs, high LMP7 expression levels were also significantly correlated with a higher histologic grade (p=0.001) and higher TIL level (p < 0.001). However, pT stage (p=0.454) and lymph node metastasis (p=0.116) were not significantly associated with LMP7 expression. LMP7 expression in tumor cells was significantly correlated with interferon-associated molecules such as HLA-ABC (p < 0.001), MxA (p < 0.001), and PKR (p < 0.001) and endoplasmic reticulum stress-associated molecules such as PERK (p=0.012), p-eIF2a (p=0.001), and XBP1 (p < 0.001). Further, LMP expression in tumor cells was significantly correlated with the cytoplasmic expression of damage-associated molecular patterns such as HMGN1 (p < 0.001) and HMGB1 (p < 0.001). These results are likewise presented in the clustering analysis (Fig. 2).
Since prognosis and adjuvant chemotherapeutic agents differ according to lymph node metastasis, we analyzed the prognostic significance of LMP7 expression in the TNBC cohort separately in groups with the presence or absence of lymph node metastasis. Patients with higher LMP7 expression showed better disease-free survival outcomes than those with no or little expression of LMP7 in the group of positive lymph node metastases (p=0.041) (Fig. 3). However, by multivariate analyses, which included histologic grade, pT stage, level of TIL, and LMP7 expression, only the TIL level (p=0.014) (Table 3) was a significantly better prognostic factor in the group. There were no differences among patients with different levels of LMP7 expression in the group without lymph node metastasis (p=0.541).
Since we only analyzed LMP7, one of three IP subunits, using immunohistochemistry, we further assessed the TCGA data to assess any correlation of IP expression and T-cell infiltration (Table 4). We also analyzed ordinary proteasome expression. The gene expression of IP subunits (PSMB8, PSMB9, and PSMB10) showed significant correlation with each other and with immune molecules (CD3D, CD3E, CD3G, and CD8), whereas the gene expression of ordinary proteasomes (PSMB5, PSMB6, and PSMB7) did not show significant association.
Cancer cells have increased proteasome levels for rapid cell proliferation. However, studies have shown that IP levels vary depending on the type of cancers [4]. Interestingly, whereas the upregulation of IP expression has been reported in prostate cancer, multiple myeloma, and lung cancer, the downregulation of IP expression has been reported in cancers of the colon [15], kidney, skin, neck, head, and esophagus [16]. Among them, upregulation of IPs is often observed in the cancer types in which the inflammatory process is important in pathogenesis of cancer. On the other hand, abnormal MHC class I expression and the loss of antigen processing are attributes of malignant cells in human and murine models that are associated with the occurrence and progression of melanoma, breast, and colorectal cancers [4]. T cell‒mediated immune tumor suppression is a complex process with numerous requirements, among which is antigen processing by the IP and presentation through HLA class I surface molecules expressed on tumor cells. The IP genes is regulated by both cell-intrinsic and cell-extrinsic factors in different types of cancer [17]. Rouette et al. [17] found that in breast cancer, the upregulation of IP genes is a cancer cell-extrinsic process correlated with the presence of interferon γ–secreting TILs. The high expression of IP genes correlates with improved survival in patients with breast cancer from TCGA data [17]. We confirmed this finding in our current study using a large number of TNBC patients who received standardized adjuvant treatments. However, LMP7 expression was not a significant prognostic factor in multivariate analysis including level of TIL. This result might be due to close association of LMP7 expression with TILs which is a strong independent prognostic factor in TNBCs.
We previously demonstrated associations between TILs, endoplasmic reticulum stress-associated molecules (PERK, p-eIF2, and XBP1), and the release of damage-associated molecular patterns (HMGB1 and HMGN1), which are associated with the activation of interferon pathway [5-8,18,19]. In the present study, we observed a significant association between TIL levels and the expression of the interferonassociated molecules MxA, PKR, HLA-ABC, and LMP7. Therefore, we can speculate that elevated endoplasmic reticulum stress induces the release of damage-associated molecular patterns to the tumor microenvironment, which activates interferon signaling in tumor cells. The elevated interferon signaling in TNBC cells then induces higher activity in the IPs, which might improve antigen presentation and result in the recruitment of TILs to the tumor tissue. We showed from our current analyses also that TNBCs with no LMP7 expression had poorer survival outcomes in those cases with lymph node metastasis, which might be caused by lack of effective antigen presentation in these tumors. However, in the consecutive breast cancer cohort, while LMP7 expression showed positive correlation with level of TILs and expression of HLA-ABC, LMP7 expression was not associated with survival in all subtypes. These results might be caused by different role of immune microenvironment in each subtype of breast cancer or by various treatment modalities given in the consecutive breast cancer cohort. Patients with this cohort had surgery for breast cancer from 1993 to 1998 and received various kinds of adjuvant treatments which were not standardized as treatment of secondary TNBC cohort.
Since proteasomes and IPs have differential preferences for cleavage sites, tumors with a high expression of IPs might have different repertoires of displayed antigens from those with a low expression of IPs. Tumor antigens that are more efficiently or poorly processed by IPs have been identified previously [20]. For example, MAGE-A3, which is a cancertestis antigen, is better processed by cells carrying IPs, while melanoma differentiation antigens such as Melan-AMART1 and gp100Pmel17 are known to be poorly processed by those cells. Although there has been no study to date that has explored which tumor antigens are more efficiently processed by IPs in breast cancer, there might be some epitopes that are better processed by IPs that are recognized by cytotoxic T lymphocytes better in breast cancers with high expression of IPs. If we could find such peptides that are recognized by the cytotoxic T lymphocyte, we could incorporate these into the design of immunotherapies such as vaccination and the adoptive transfer of T lymphocytes reactive to the epitopes.
The limitations of this study included its retrospective design and the evaluation of LMP7 alone among IP subunits. Further studies should examine other IP subunits and their relationships, and identify tumor epitopes that are better processed by IPs.
ACKNOWLEDGMENTS
This study was supported by the Korean Health Technology R&D Project, Ministry of Health & Welfare (HI17C0337).
References
1. van Rooijen JM, Stutvoet TS, Schroder CP, de Vries EG. Immunotherapeutic options on the horizon in breast cancer treatment. Pharmacol Ther. 2015; 156:90–101.

2. Kast J. Immunoproteasome deficiency in non-small cell lung cancer and its relevance to immunotherapy. J Thorac Dis. 2016; 8:E1082–6.

3. Shashova EE, Lyupina YV, Glushchenko SA, Slonimskaya EM, Savenkova OV, Kulikov AM, et al. Proteasome functioning in breast cancer: connection with clinical-pathological factors. PLoS One. 2014; 9:e109933.

4. Kaur G, Batra S. Emerging role of immunoproteasomes in pathophysiology. Immunol Cell Biol. 2016; 94:812–20.

5. Kim JY, Heo SH, Song IH, Park IA, Kim YA, Gong G, et al. Activation of the PERK-eIF2alpha pathway is associated with tumor-infiltrating lymphocytes in HER2-positive breast cancer. Anticancer Res. 2016; 36:2705–11.
6. Lee HJ, Park IA, Song IH, Shin SJ, Kim JY, Yu JH, et al. Tertiary lymphoid structures: prognostic significance and relationship with tumour-infiltrating lymphocytes in triple-negative breast cancer. J Clin Pathol. 2016; 69:422–30.

7. Lee HJ, Song IH, Park IA, Heo SH, Kim YA, Ahn JH, et al. Differential expression of major histocompatibility complex class I in subtypes of breast cancer is associated with estrogen receptor and interferon signaling. Oncotarget. 2016; 7:30119–32.

8. Park IA, Heo SH, Song IH, Kim YA, Park HS, Bang WS, et al. Endoplasmic reticulum stress induces secretion of high-mobility group proteins and is associated with tumor-infiltrating lymphocytes in triple-negative breast cancer. Oncotarget. 2016; 7:59957–64.

9. Kim YA, Lee HJ, Heo SH, Park HS, Park SY, Bang W, et al. MxA expression is associated with tumor-infiltrating lymphocytes and is a prognostic factor in triple-negative breast cancer. Breast Cancer Res Treat. 2016; 156:597–606.

10. Lee HJ, Seo AN, Park SY, Kim JY, Park JY, Yu JH, et al. Low prognostic implication of fibroblast growth factor family activation in triple-negative breast cancer subsets. Ann Surg Oncol. 2014; 21:1561–8.

11. Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version). Arch Pathol Lab Med. 2010; 134:e48–72.
12. Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch Pathol Lab Med. 2014; 138:241–56.

13. Torigoe T, Asanuma H, Nakazawa E, Tamura Y, Hirohashi Y, Yamamoto E, et al. Establishment of a monoclonal anti-pan HLA class I antibody suitable for immunostaining of formalin-fixed tissue: unusually high frequency of down-regulation in breast cancer tissues. Pathol Int. 2012; 62:303–8.

14. Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012; 490:61–70.
15. Cabrera CM, Jimenez P, Cabrera T, Esparza C, Ruiz-Cabello F, Garrido F. Total loss of MHC class I in colorectal tumors can be explained by two molecular pathways: beta2-microglobulin inactivation in MSI-positive tumors and LMP7/TAP2 downregulation in MSI-negative tumors. Tissue Antigens. 2003; 61:211–9.
16. Miller Z, Ao L, Kim KB, Lee W. Inhibitors of the immunoproteasome: current status and future directions. Curr Pharm Des. 2013; 19:4140–51.

17. Rouette A, Trofimov A, Haberl D, Boucher G, Lavallee VP, D'Angelo G, et al. Expression of immunoproteasome genes is regulated by cell-intrinsic and -extrinsic factors in human cancers. Sci Rep. 2016; 6:34019.

18. Lee HJ, Seo JY, Ahn JH, Ahn SH, Gong G. Tumor-associated lymphocytes predict response to neoadjuvant chemotherapy in breast cancer patients. J Breast Cancer. 2013; 16:32–9.

Fig. 1.
Immunohistochemical staining of LMP7 in breast cancer tissues. Representative images are shown (×200).
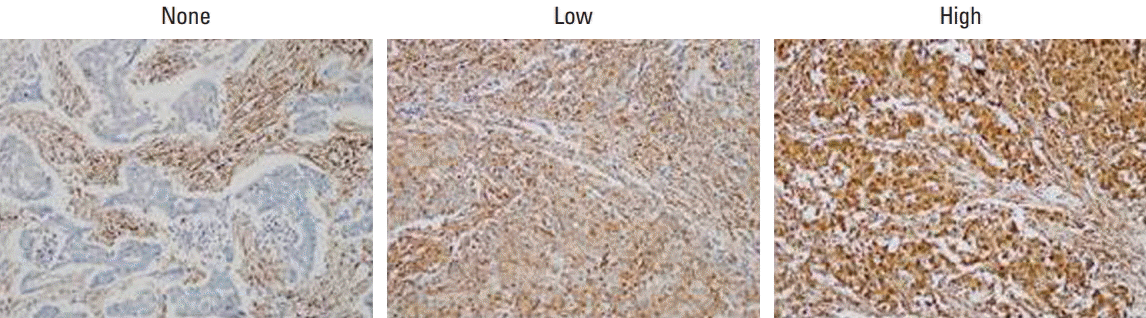
Fig. 2.
Heat map plot for LMP7 expression with interferon-associated molecules, endoplasmic reticulum stress-associated molecules, damage-associated molecular patterns, and amount of tumor-infiltrating lymphocytes.
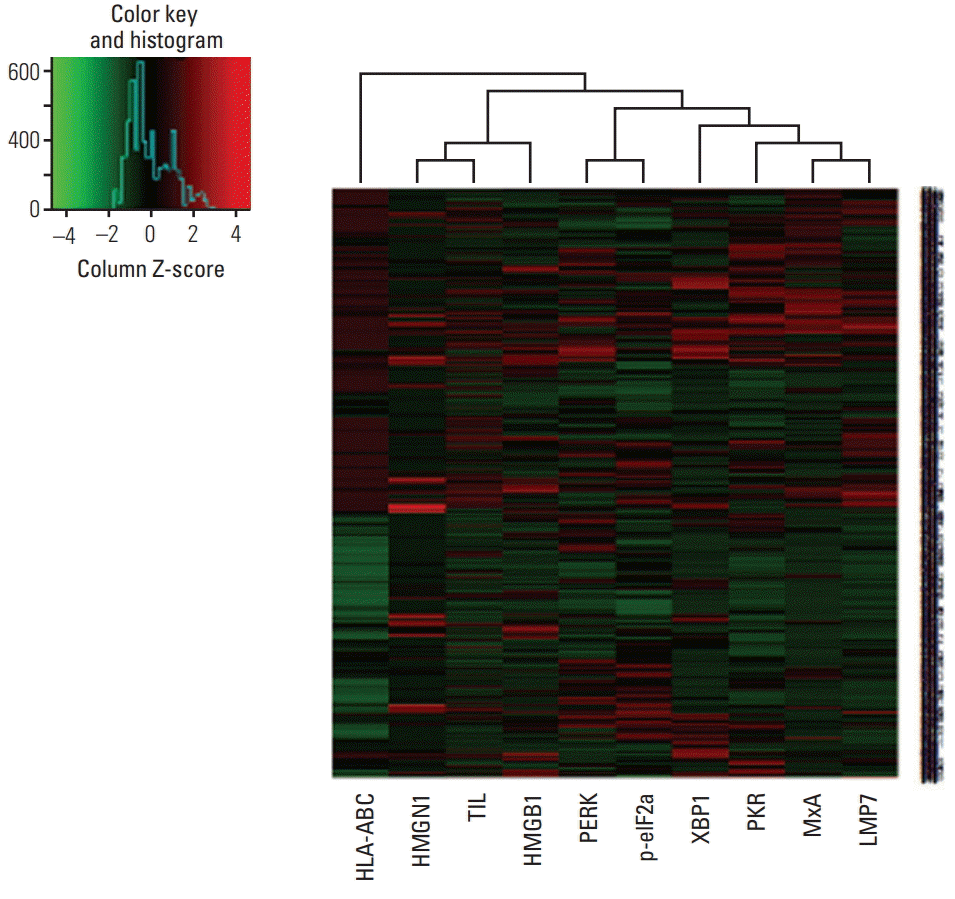
Fig. 3.
Relationship between LMP7 expression and disease-free survival in the triple-negative breast cancer cohort. Kaplan-Meier survival curves are shown for disease-free survival in the group of positive lymph node (LN) metastases and in the group of negative LN metastases.
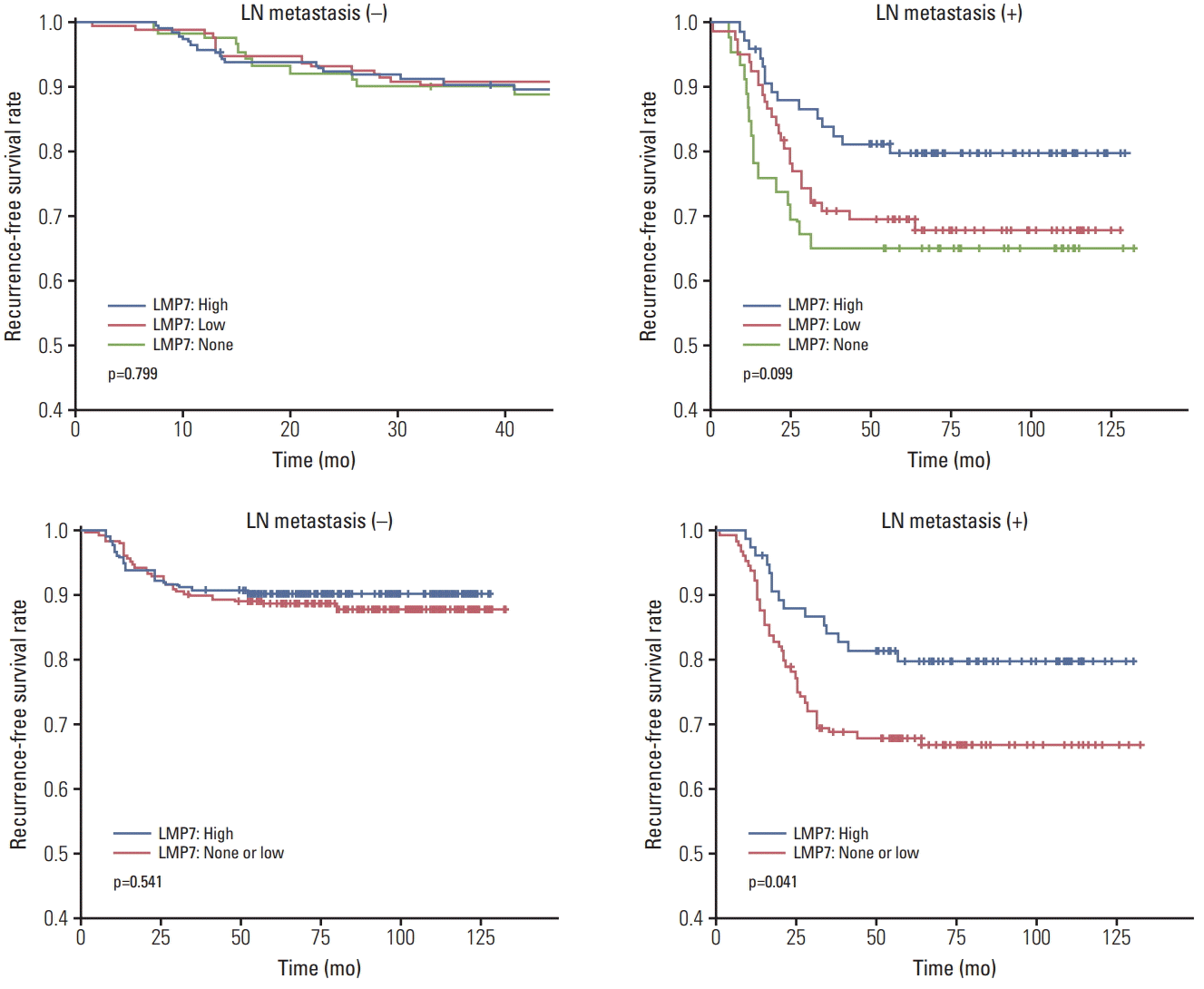
Table 1.
Comparison of pathological variables according to LMP7 expression in the first consecutive breast cancer cohort
Table 2.
Comparison of pathological variables and various immune-related markers according to LMP7 expression in the second TNBC cohort
Table 3.
Univariate and multivariate disease-free survival analyses in positive lymph node metastases group of TNBC cohort
Table 4.
Correlations between the gene expression levels among the proteasome, IP, and T-cell markers




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download