This article has been retracted. See "RETRACTION: Circular RNA-ZFR Inhibited Cell Proliferation and Promoted Apoptosis in Gastric Cancer by Sponging miR-130a/miR-107 and Modulating PTEN " in Volume 56 on page 697.
Abstract
Purpose
This study aimed to probe into the associations among circular RNA ZFR (circ-ZFR), miR-130a/miR-107, and PTEN, and to investigate the regulatory mechanism of circ-ZFR‒miR-130a/miR-107‒PTEN axis in gastric cancer (GC).
Materials and Methods
GSE89143 microarray data used in the study were acquired from publicly available Gene Expression Omnibus database to identify differentially expressed circular RNAs inGC tissues. The expressions of circ-ZFR, miR-130a, miR-107, and PTEN were examined by real-time reverse transcription polymerase chain reaction, while PTEN protein expression was measured by western blot. The variation of GC cell proliferation and apoptosis was confirmed by cell counting kit-8 assay and flow cytometry analysis. The targeted relationships among circZFR, miR-130a/miR-107, and PTEN were predicted via bioinformatics analysis and demonstrated by dual-luciferase reporter assay and RNA immunoprecipitation assay. The impact of ZFR on gastric tumor was further verified in xenograft mice model experiment.
Results
Circ-ZFR and PTEN were low-expressed whereas miR-107 and miR-130a were highexpressed in GC tissues and cells. There existed targeted relationships and interactions between miR-130a/miR-107 and ZFR/PTEN. Circ-ZFR inhibited GC cell propagation, cell cycle and promoted apoptosis by sponging miR-107/miR-130a, while miR-107/miR-130a promoted GC cell propagation and impeded apoptosis through targeting PTEN. Circ-ZFR inhibited cell proliferation and facilitated apoptosis in GC by sponging miR-130a/miR-107 and modulating PTEN. Circ-ZFR curbed GC tumor growth and affected p53 protein expression in vivo.
Gastric cancer (GC) is one of the most common lethal neoplasms and also the second cause of cancer-incurred death around the world [1]. In the recent years, with rapid advances of diagnostic apparatus and therapeutic approaches, the mortality and morbidity rates of GC have been on a steady downward trend [2]. However, the prognostic outcome of advanced stage disease still remains poor and 5-year overall survival rate is less than 30% due to frequent tumor recurrence and metastasis [3]. Therefore, it is imperative to investigate the in-depth molecular mechanism of GC for the development of effective targeted therapies.
Circular RNAs (circRNAs) are a large class of endogenously expressed non-coding RNAs characterized by covalently closed loop structures with neither 5′ to 3′ polarity nor polyadenylated tail [4]. Numerous researches have revealed that circRNAs play critical roles in regulating multiple cellular activities and pathological processes [5]. It has been also substantiated that circRNAs participate in the initiation and progression of several types of cancers [6]. Huang et al. [7] reported that circular RNA 0000745 (hsa_circ_0000745) expression was downregulated that is closely correlated with tumor differentiation. Guo et al. [8] disclosed that overexpression of circular RNA 0000069 (hsa_circ_0000069) promoted cell propagation and metastasis in colorectal cancer. Zhang et al. [9] verified that increased circ-UBAP2 could facilitate cell growth and inhibit apoptosis in osteosarcoma. Nonetheless, the regulatory function of circular RNA ZFR (circ-ZFR) on GC remains unknown, which led us to wonder whether or not circ-ZFR could serve as a novel biomarker for GC diagnosis and treatment.
MicroRNAs (miRNAs) are small non-coding RNAs that bind to the 3′-untranslated regions (3′-UTR) of target mRNAs and involved in the several types of cancer as tumor facilitator or inhibitor, thereby potentially modulating the biological processes [10,11]. Furthermore, circRNAs have been demonstrated to function as miRNA sponge to regulate gene expression in multiple cancers [12]. For instance, circular RNA MTO1 was found to suppress hepatocellular carcinoma progression by sponging miR-9 in the recent study of Han et al. [13]. Li et al. [14] demonstrated that circular RNA FUT10 reduced proliferation and facilitated differentiation of myoblasts by sponging miR-133a. Nevertheless, fewer studies focused on the effects of circ-ZFR on GC, and thus the regulatory function of circ-ZFR‒miRNAs‒mRNAs axis in GC needs to be further elucidated.
Herein, we were dedicated to investigating the associations among circ-ZFR, miR-130a/miR-107, and PTEN as well as the regulatory mechanism of circ-ZFR‒miR-130a/miR-107–PTEN axis in GC.
GC tissues and adjacent tissue samples were collected from 48 GC patients in the Gastroenterology Center of First Affiliated Hospital of Kunming Medical University from February 2011 to February 2016. The final diagnosis of each patient was confirmed by histopathology and evaluated in accordance with tumor nodes metastasis (TNM) staging system and National Comprehensive Cancer Network Oncology Clinical Practice Guidance (V.1.2012).
Total RNA expression was analyzed through Affymetrix Human Genome U133 Plus 2.0 Array (Affymetrix, Santa Clara, CA). Robust multiarray average normalization was performed using R language and environment (http://www.r-project.org/). GSE89143 microarray data used in the study were acquired from publicly available Gene Expression Omnibus database to identify differentially expressed circRNAs. The screening threshold of differentially expressed circRNAs was set as log(fold change) > 2 and p < 0.05.
Human gastric epithelial cell line GES-1 and human GC cell lines AGS, AZ521, and HGC-27 were acquired from BeNa Culture Collection (BNCC, Beijing, China). All cell lines received short tandem repeat profiling authentication and mycoplasma contamination tests. GC cell line AGS was cultured in 90% Ham’s F12 nutrient medium (Sigma, St. Louis, MO) supplemented with 10% fetal bovine serum (FBS); AZ521 cells were stored in Dulbecco’s modified Eagle medium plus 10% FBS (Gibco, Grand Island, NY); the culture of HGC-27 cell line was performed in 80% RPMI-1640 (Gibco) containing 20% FBS. All the cell lines were maintained in the medium at 37°C overnight.
AGS cells in the logarithmic growth phase were harvested and 2×105 cells were inoculated onto 6-well plates. Subsequently, AGS cells were respectively transfected with pcDNA-3.1‒circ-ZFR or pcDNA3.1-PTEN, miR-107 mimics or inhibitor, miR-130a mimics or inhibitor following the instructions of Lipofectamine 3000 reagent (Life Technologies, Carlsbad, CA) for 24 hours. The experiment grouping was as following: (1) negative control (NC) group and pcDNA3.1-ZFR group; (2) NC group, pcDNA3.1-ZFR group, miR-107‒mimics group, miR-107‒inhibitor group, and miR-107–mimics+pcDNA3.1-ZFR group; (3) NC group, miR-130a–mimics group, miR-130a‒inhibitor group, miR-130a–mimics+pcDNA3.1-ZFR group; (4) NC group, miR-107–mimics group, pcDNA3.1-PTEN group, and pcDNA3.1-PTEN+miR-107–mimics group; (5) NC group, miR-130a–mimics group, pcDNA3.1-PTEN group, and pcDNA3.1-PTEN+miR-130a–mimics group.
The nuclear and cytoplasmic extracts were prepared through NE-PER nuclear and cytoplasmic extraction reagents (Pierce, Brockville, ON, Canada), followed by total RNA isolation using TRIzol (Life Technologies). In terms of RNase R treatment, total RNA (2 mg) with or without 3 U/mg of RNase R was incubated for 30 minutes at 37°C (Epicentre Technologies, Madison,WI). Subsequently, RNeasy MinElute cleaning Kit (Qiagen, Hilden, Germany) was utilized to purify the resulting RNA. TaqMan MicroRNA assays (Life Technologies) was employed to quantify the amount of mature miRNA, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as an internal standard. To quantify the amount of mRNA and circRNA, cDNA was synthesized from 500 ng of RNA by via the PrimeScript RT Master Mix (Takara, Shiga, Japan), and the real-time polymerase chain reaction (PCR) results were analyzed through SYBR Premix Ex Taq II (Takara Bio Inc., Otsu, Japan). To confirm the absolute quantity of RNA, the purified PCR product, which is amplified from cDNA in line with the circ-ZFR sequence, was serially diluted to produce a standard curve. The primer sequences were exhibited in Table 1.
Total protein was extracted using RIPA buffer (Sigma). The protein concentration was verified by the BCA protein assay kit (Pierce). The proteins were segregated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (Bio-Rad, Hercules, CA) and transferred onto polyvinylidene difluoride membrane. The primary antibodies used were anti-PTEN (1:1,000, ab170941, Abcam, Cambridge, MA), anti-p53 (1:1,000, ab131442, Abcam), and anti-GAPDH (1:10,000, ab181602, Abcam). The membranes were washed and then incubated in goat anti-rabbit horseradish peroxidase‒conjugated secondary antibody (1:2,000) for 2 hours at 37°C. GAPDH was deemed an internal control. An enhanced chemiluminescence kit (Life Technology) used to visualize the immunoblot bands, of which the optical density was analyzed by ImageJ2X software.
Nude mice tumor tissues were fixed by paraformaldehyde solution, dewaxed in xylene, rinsed with phosphate buffered saline (PBS) and incubated with 3% hydrogen peroxide in 50% methanol for 30 minutes at 37°C. After the elimination of endogenous peroxidase activity, the sections were rinsed in PBS again and incubated in protein block solution (Bio-Genex, San Ramon, CA) in a humid chamber for 30 minutes. One hundred microliters PTEN was added to the sections, which were then incubated with primary antibody anti-ZFR (1:100, ab170941, Abcam) in a humid chamber overnight. Sections exposed to diluents alone without primary antibody served as negative controls. The slides were then rinsed three times in PBS for 5 minutes each and incubated with goat antimouse secondary antibody for 30 minutes. The reaction was developed using the peroxidase substrate diaminobenzidine. The sections were counterstained with hematoxylin.
Cell proliferation was assessed using cell counting kit-8 (CCK-8) kit (Dojindo Laboratories, Kumamoto, Japan). AGS cells were seeded in the 96-well plates (2×103 cells/well) containing 10 μL CCK-8 solutions (Beyotime, Jiangsu, China) and were incubated at 37°C for 1 hour. The absorbance was measured by microplate reader (Pharmacia Biotech, Piscataway, NJ) at 450 nm.
Cell cycle and apoptosis of transfected cells were determined by flow cytometry assay. After transfection for 96 hours, 1×106 cells were digested using 0.25% trypsin without EDTA, washed by pre-cooled PBS for three times and collected by centrifugation at 2,000 rpm. Then cells were washed twice using 5% bovine serum albumin (BSA), after which the supernatant was discarded and then the cells were added with 300 μL 5% BSA and 700 μL of 70% pre-cooled alcohol and stored in a fridge at –20°C overnight. Subsequently, the cells were centrifuged at 2,000 rpm for 5 minutes, rinsed and resuspended in PBS. And then, AGS cells were incubated with 1 μL 10 mg/μL RnaseA for 20 minutes. Three hundred microliters 10 μg/mL propidum iodide (PI; Sigma) was applied to cell staining for 15 minute at 4°C. Furthermore, cell apoptosis was performed by reference to Annexin V-FITC apoptosis detection kit instructions (Sigma), 100 μL binding buffer and 5 μL Annexin-V-FITC (20 μg/mL) were added into each tube, followed by 15-minute incubation in the dark. Next, 150 μL binding buffer and 10 μL PI dye (50 μg/mL) were added into tube. Cell cycle and apoptosis rate was measured by FACSCanto II flow cytometer (BD Biosciences, San Jose, CA).
RNA immunoprecipitation (RIP) assay was conducted via the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA) following the producer’s protocol. Briefly, 1×107 cells at 80% confluence were harvested and were lysed in RIP lysis buffer. After that, 100 μL cell lysate was incubated with RIP buffer containing magnetic beads conjugated with human anti-argonaute 2 antibody (Millipore) or NC normal mouse IgG (Millipore). The specimens were incubated with proteinase K to segregate immunoprecipitated RNA. The extracted RNAs were further used to confirm circRNA-MYLK through real-time reverse transcription polymerase chain reaction (qRT-PCR).
The relationship between circ-ZFR and miR-130a/miR-107 was verified through starbase prediction software (http://starbase.sysu.edu.cn/) and the relationship between miR-130a/miR-107 and PTEN was detected by TargetScan (http://www.targetscan.org/mamm_31/). Cells were seeded into 96-well plates (100 μL/well) and incubated at 37°C overnight. Plasmid vector pcDNA3.1 (0.2 μg) and FugeneHD (0.3 μL) were purchased from Promega (Beijing, China). Wild type and mutated type ZFR‒3′-UTR were respectively inserted into the XhoI/NotI sites of pcDNA3.1 to construct recombinant. Afterwards, the cells were co-transfected with recombinant pcDNA3.1-ZFR, miR-107 mimics, miR-107 inhibitor, miR-130a mimics, miR-130a inhibitor, or miR-NC using Lipofectamine 2000. At 48 hours for transfection, the relative luciferase activity was confirmed following the Dual-Luciferase Reporter Assay kit instructions (Promega). Firefly luciferase activity was normalized relative to Renilla luciferase.
Ten male BALB/c Nude mice (8-week-old) were obtained from Centre of First Affiliated Hospital of Kunming Medical University and used for the construction of nude mice model. All mice were kept under the pathogen-free circumstance during the entire research. Generally, 1×107 AGS cells transfected with pcDNA 3.1-ZFR overexpression plasmids were subcutaneously injected into the flank of nude mice. All mice were sacrificed at the fifth week after treatment. Tumor volume was measured following the formula of “tumor volume=length×width×width/2.”
Statistical data were analyzed using GraphPad Prism 6.0 (GraphPad Software, San Diego, CA) and documented as means±standard deviation. The Pearson’s correlation coefficient analysis was employed to confirm the correlations. Comparisons among groups were assessed through Student's t test and one-way ANOVA. p < 0.05 was deemed statistically significant.
This animal experiment has been conducted following the authenticated animal protocols of Ethical Committee of Animal Welfare of First Affiliated Hospital of Kunming Medical University.
This research had gotten ratification from the Human Research Ethics Committee of First Affiliated Hospital of Kunming Medical University and obtained informal written consents from all subjects. All clinical specimens and data were collected in a randomized and double-blind way throughout the research.
To explore the role of circRNAs in the development of gastric cancer, we analyzed GC tumor tissues and its matched adjacent tissues by using microarrays. The different expression circRNAs were shown in Fig. 1A. We screened the top eight high expressions and low expressions circRNAs in GC and graphing the Heatmap, and then circ-ZFR was decreased in GC tissue and selected for further study (Fig. 1B). Meanwhile, qRT-PCR also displayed that circ-ZFR was drastically downregulated in tumor tissues compared with adjacent tissues (p < 0.05) (Fig. 1C). Moreover, the expression of circ-ZFR was considerably lower in human GC cell lines (AGS, AZ521, and HGC-27) than in gastric epithelial cell line GES-1 (p < 0.05) (Fig. 1D). Since AGS cell line presented the most significant difference on circ-ZFR expression in comparison with GES-1 cell line (p < 0.01), it was selected for the later experiments. In addition, to further confirm the characteristics of circ-ZFR, we used a highly processive 3′ to 5′ exoribonuclease (RNase R enzyme) that does not act on circRNAs but linear RNAs. As expected, circ-ZFR was resistant to RNase treatment in contrast to GAPDH as shown in Fig. 1E.
qRT-PCR results presented that circ-ZFR expression was remarkably increased after transfection with pcDNA3.1-ZFR (p < 0.01) (Fig. 2A). Furthermore, overexpression of circ-ZFR observably suppressed GC cell proliferation, as confirmed in CCK-8 assay (p < 0.05) (Fig. 2B). In addition, flow cytometry analysis disclosed the variance of cell cycle and apoptosis condition after transfection with pcDNA3.1-ZFR. As exhibited in Fig. 2C and D, the percentage of cells arrested in G0/G1 phase was significantly higher in pcDNA3.1-ZFR group than in NC group, while that of arrested cells in S phase relatively declined compared with NC group (both p < 0.01). Meanwhile, the apoptosis rate of GC cells significantly increased after transfection (p < 0.05). Overall, overexpression of circ-ZFR arrested cell propagation and cell cycle and promoted apoptosis in GC.
Through starbase, a bioinformatics prediction website, we entered circ-ZFR into starbase and search the potential targeting miRNAs, founding that the binding sites of miR-107/miR-130a on 3′-UTR of ZFR were showed in Fig. 3A and B. Depending on DIANA TOOLS-mirPath v.3 website, we sought to find a pathway that is co-influenced by miR-130a and miR-107, p53 signaling pathway was selected for further study (Fig. 3C). And then, we crossed the targeting gene was regulated by both p53 pathway and miR-130a/miR-107 via venn diagram, and intersection was PTEN showed in Fig. 3D. Finally, we utilized Targetscan to find out the relationship between miR-130a/miR-107 and PTEN (Fig. 3E). The network clarified the mechanism of circ-ZFR, miR-130a/miR-107, and PTEN in GC showed in Fig. 3F, which the experimental research idea was depended on.
qRT-PCR displayed that miR-107 mimics and miR-130a mimics contributed to miR-107 and miR-130a overexpression respectively (p < 0.05) (Fig. 4A). While, the expression of circ-ZFR was detected and the results revealed that miR-107 mimics and miR-130a mimics have little influences on circ-ZFR level (Fig. 4B). That is to say, the relationship between circ-ZFR and miR-107/miR-130a was unidirectional. Moreover, overexpression of miR-107 and miR-130a both significantly repressed the luciferase activity of the luciferase reporter containing circ-ZFR 3′-UTR‒WT but not the reporter containing circ-ZFR 3′-UTR‒Mut, as shown in dual luci-ferase reporter assay (both p < 0.05) (Fig. 4C and D). RIP assay verified that there existed some interactions between circ-ZFR and miR-107/miR-130a both (p < 0.05) (Fig. 4E and F). Furthermore, we also verified that miR-107/miR-130a could bind to 3’ UTR of PTEN (both p < 0.05) (Fig. 4G and H). Additionally, in Fig. 5A and B, miR-107 and miR-130a were found to be highly expressed, whereas PTEN was lowly expressed in tumor tissues compared with adjacent tissues (p < 0.05). Furthermore, Pearson correlation analysis revealed that the expressions of miR-107 and miR-130a were both negatively correlated with those of ZFR and PTEN (Fig. 5C and F).
qRT-PCR displayed that miR-107/miR-130a mimics led to overexpression of miR-107/miR-130a, while miR-107/miR130a inhibitor downregulated miR-107/miR-130a expressions (p < 0.05) (Fig. 6A and C). CCK-8 assay (Fig. 6B and D) revealed that upregulation of miR-107 and miR-130a expression both promoted cell propagation, whereas downregulation of miR-107 and miR-130a expressions restrained propagation compared with NC group (p < 0.05). Moreover, no significant disctinction was found between miR-107/miR130a+pcDNA3.1-ZFR group and NC group (p > 0.05). However, the proliferation ability of the cells in miR-107/miR-130a +pcDNA3.1-ZFR group was significantly stronger than that in pcDNA3.1-ZFR group, while weaker compared with miR107/miR-130a–mimics group (p < 0.05).
For the analysis of cell cycle and apoptosis, flow cytometry assay (Fig. 7A and B) indicated that the proportion of the cells arrested in G1 phase in pcDNA3.1-ZFR group, miR-107 inhibitor group and miR-130a inhibitor group was noticeably larger than in NC group. Conversely, the arrested cell in G1 phase in miR-107–mimics group and miR-130a–mimics group was relatively fewer (p < 0.05). Nonetheless, the arrested cell in G1 phase in miR-107/miR-130a+pcDNA3.1-ZFR group was remarkably fewer than that in pcDNA3.1-ZFR group, while more than that in miR-107/miR-130a–mimics group (p < 0.05). Similarly, no conspicuous distinction was found between miR-107/miR-130a+pcDNA3.1-ZFR group and NC group (p > 0.05). The apoptosis of cells transfected with pcDNA3.1-ZFR and miR-107/miR-130a inhibitor was significantly enhanced, while that of the cells transfected with miR-107/miR-130a mimics was remarkably inhibited (p < 0.05) (Fig. 8A and B). No significant difference of apoptosis rate was observed between miR-107/miR 130a+pcDNA-3.1-ZFR group and NC group (p > 0.05). However, the cell apoptosis rate in miR-107/miR-130a+pcDNA3.1-ZFR group was observably lower than that in pcDNA3.1-ZFR group, while significantly higher compared with miR-107/miR-130a–mimics group (p < 0.05).
Western blot confirmed that the expression of PTEN was remarkably repressed after transfection with miR-107–mimics/miR-130a–mimics (both p < 0.05), while it was overexpressed followed transfection with pcDNA3.1-ZFR and miR-107 inhibitor/miR-130a inhibitor (both p < 0.01). In addition, no significant difference was found between miR-107/miR-130a+pcDNA3.1-ZFR group and NC group (p > 0.05) (Fig. 9A and B). qRT-PCR results displayed that after transfection with pcDNA3.1-PTEN, the expression of pcDNA-3.1-PTEN in GC cells was dramatically upregulated (p < 0.01) (Fig. 9C). Furthermore, CCK-8 assay exhibited that overexpression of PTEN significantly attenuated GC cell propagation ability, while that of miR-107/miR-130a mimics promoted cell propagation. Meanwhile, the proliferation ability of the cells in miR-107/miR-130a‒mimics+pcDNA3.1-PTEN group was significantly stronger than that in pcDNA3.1-PTEN group, while weaker compared with miR-107/miR-130a‒mimics group (p < 0.05) (Fig. 9D and E). No obvious distinction of propagation ability was found between miR-107/miR-130a‒mimics+pcDNA3.1-PTEN group and NC group (p > 0.05).
Additionally, flow cytometry assay (Fig. 10A and B) indicated that the proportion of the cells arrested in G1 phase in miR-107/miR-130a–mimics group dramatically reduced, while that in S phase increased. Conversely, the arrested cells in G1 phase in pcDNA3.1-PTEN group conspicuously increased in comparison with NC group, whereas those in S phase relatively decreased (p < 0.05). Nonetheless, the percentage of arrested cell in G1 phase in miR-107/miR-130a+pcDNA3.1-PTEN group was significantly smaller than that in pcDNA3.1-PTEN group, while larger than that in miR-107/miR-130a–mimics group (p < 0.05). Furthermore, the apoptotic rate of the cells transfected with miR-107/miR-130a mimics considerably fell, while cell apoptosis drastically rose in pcDNA3.1-PTEN group (p < 0.05) (Fig. 11A and B). No significant difference of apoptosis ratio was detected between miR-107/miR-130a+pcDNA3.1-PTEN group and NC group (p > 0.05). Nevertheless, the cell apoptosis rate in miR-107/miR-130a+pcDNA3.1-PTEN group was considerably lower than that in pcDNA3.1-PTEN group, while significantly higher compared with miR-107/miR-130a–mimics group (p < 0.05).
As displayed in Fig. 12A, the tumor volume of the mice in pcDNA3.1-ZFR group grew more slowly than that in NC group (p < 0.05). Moreover, the tumor weight of mice in pcDNA3.1-ZFR group was significantly lighter compared with NC group (p < 0.05) (Fig. 12B and C). The expression level of miR-107 and miR-130a was decreased compared with NC group after transfected with pcDNA3.1-ZFR detected by qRT-PCR (p < 0.01) (Fig. 12D and E). Meanwhile, IHC assay revealed that after transfection with pcDNA3.1-ZFR, the expression level of p53 and PTEN protein dramatically increased in comparison with NC group (p < 0.01) (Fig. 12F and G).
Based on a series of experiment, we verified the regulatory role of circ-ZFR–miR-130a/miR-107–PTEN network in GC. We first identified that circ-ZFR was downregulated in GC and acted as a tumor inhibitor in cell propagation and apoptosis of GC. Meanwhile, we validated the targeted relationships between miR-107/miR-130a and PTEN/circ-ZFR. Furthermore, we disclosed that circ-ZFR expression was negatively correlated with miR-130a and miR-107 expressions, which were also correlated inversely with their target PTEN expression. Our study finally demonstrated that circ-ZFR inhibited GC cell proliferation and promoted apoptosis by sponging miR-130a/miR-107 and modulating PTEN.
CircRNAs, a novel type of endogenous non-coding RNAs, have been recently considered a crucial regulator of gene expression and pathological processes. Accumulating researches have suggested that aberrant circRNA expression plays important roles in carcinogenesis and tumor progression [15]. Fu et al. [16] demonstrated that circular RNA 0004018 (hsa_circ_0004018) was lowly expressed and played a role in carcinogenesis and metastasis of hepatocellular carcinoma. In the recent study of Li et al. [17] circular RNA 0000096 (hsa_circ_0000096) was also found to be significantly downregulated in GC tissues and knockdown of hsa_circ_0000096 significantly inhibited cell propagation and migration in vitro and in vivo. Herein, circ-ZFR was identified to be observably downregulated in GC tissues based on microarray analysis, and overexpression of circ-ZFR could retard propagation and cell cycle and promoted apoptosis in GC confirmed by CCK-8 and apoptosis assay.
It is well recognized that miRNAs play a critical role in the regulation of gene expression and various biological processes, such as proliferation, metastasis, apoptosis and so forth. The circRNA-miRNA association and their interaction influence on various cancers have been widely studied. CircRNAs has been demonstrated to serve as miRNA sponge, thereby regulating gene transcription and cellular activities in many researches [18]. Liang et al. [19] disclosed that circ-ABCB10 promoted breast cancer proliferation and progression through sponging miR-1271. Tang et al. [20] reported that circ_0001982 facilitated breast cancer cell carcinogenesis through repressing miR-143. Zhong et al. [21] unraveled that circ-TCF25 accelerated cell propagation and mobility in bladder cancer through sponging miR-103a-3p/miR-107. However, Zou et al. [22] found that miR-107 was a tumor progression promoter in hepatocellular carcinoma, which suggested that miR-107 takes different effects in different cancers. A recent study also revealed that miR-107 was upregulated in GC [23], which displayed the similar result as our study. In the current study, we predicted the association between circ-ZFR and miR-130a-3p/miR-107, and substantiated the circ-ZFR‒miR-130a-3p/miR-107 regulatory loop in GC. Circ-ZFR inhibited GC cell propagation, induced cell cycle arrest and promoted apoptosis by sponging miR-107/miR-130a.
PTEN, as a tumor suppressor, is mutated in a large number of cancers at high frequency [24]. It negatively regulates intracellular levels of phosiphatidylinositol-3,4,5-trisphosphate in cells and AKT/PKB signaling pathway, suppressing cellular functional activities [25]. Actually, emerging evidence has substantiated that PTEN could be targeted and regulated by miR-130a and miR-107 to influence cancerous cellular activities [26,27]. Nonetheless, there are fewer studies on the targeted relationships between miR-130a/miR-107 and PTEN in GC. Herein, we verified that miR-130a/miR-107 could target and modulate PTEN expression through bioinformatics analysis, dual-luciferase reporter gene assay and RIP assay.
In addition, circRNAs have been demonstrated to act as miRNA sponges and play crucial roles in regulating gene expression through a circRNA-miRNA-gene pathway. Chen et al. [28] substantiated that circ-WDR77 affected vascular smooth muscle cells proliferation and migration by sponging miR-124 and modulating its target FGF-2. Peng et al. [29] revealed that circ-ZNF609 regulated AKT3 expression by sponging miR-150-5p in Hirschsprung disease. Xie et al. [30] found that circRNA-001569 exerted influence on the propagation and invasiveness of colorectal cancer via circRNA-001569‒miR-145-E2F5/BAG4/FMNL2 network. In our study, circ-ZFR was identified as a sponge of miR-130a/miR-107 and indirectly modulated their target PTEN expression, thereby repressing GC cell propagation and promoting apoptosis (Fig. 13). There were still some shortcomings in our research. Only the effect of circ-ZFR was explored in vivo, while the effect of miR-130a and miR-107 in vivo still remained unclear. Further study ought to be put into practice. On the other hand, we only focus on the influences of circ-ZFR–miR-130a/miR-107–PTEN axis in gastric cancer. In the follow-up study, we will pay attention to other molecular mechanisms and signaling pathways on GC procession.
In conclusion, circ-ZFR inhibited cell proliferation and promoted apoptosis in GC by sponging miR-130a/miR-107 and modulating PTEN. The current research revealed a novel molecular mechanism of circ-ZFR in regulating GC cell propagation, cell cycle and apoptosis, which provided a comprehensive insight into the regulatory role of circ-ZFR–miR-130a/miR-107–PTEN axis in GC and facilitated the discovery of novel therapeutic targets in GC treatment.
ACKNOWLEDGMENTS
This study was supported by Leader Training Program in Medical Subjects of Health and Family Planning commission of Yunnan Province (D-201657).
References
1. Xi HQ, Zhang KC, Li JY, Cui JX, Gao YH, Wei B, et al. RNAimediated inhibition of Lgr5 leads to decreased angiogenesis in gastric cancer. Oncotarget. 2017; 8:31581–91.

2. Li X, Li H, Zhang R, Liu J, Liu J. MicroRNA-449a inhibits proliferation and induces apoptosis by directly repressing E2F3 in gastric cancer. Cell Physiol Biochem. 2015; 35:2033–42.

3. Allemani C, Weir HK, Carreira H, Harewood R, Spika D, Wang XS, et al. Global surveillance of cancer survival 1995-2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet. 2015; 385:977–1010.

4. Liu J, Liu T, Wang X, He A. Circles reshaping the RNA world: from waste to treasure. Mol Cancer. 2017; 16:58.

6. Fu L, Wu S, Yao T, Chen Q, Xie Y, Ying S, et al. Decreased expression of hsa_circ_0003570 in hepatocellular carcinoma and its clinical significance. J Clin Lab Anal. 2017 May 11 [Epub]. https://doi.org/10.1002/jcla.22239.

7. Huang M, He YR, Liang LC, Huang Q, Zhu ZQ. Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World J Gastroenterol. 2017; 23:6330–8.

8. Guo JN, Li J, Zhu CL, Feng WT, Shao JX, Wan L, et al. Comprehensive profile of differentially expressed circular RNAs reveals that hsa_circ_0000069 is upregulated and promotes cell proliferation, migration, and invasion in colorectal cancer. Onco Targets Ther. 2016; 9:7451–8.

9. Zhang H, Wang G, Ding C, Liu P, Wang R, Ding W, et al. Increased circular RNA UBAP2 acts as a sponge of miR-143 to promote osteosarcoma progression. Oncotarget. 2017; 8:61687–97.

10. Izaurralde E. GENE REGULATION. Breakers and blockersmiRNAs at work. Science. 2015; 349:380–2.
11. Gattolliat CH, Uguen A, Pesson M, Trillet K, Simon B, Doucet L, et al. MicroRNA and targeted mRNA expression profiling analysis in human colorectal adenomas and adenocarcinomas. Eur J Cancer. 2015; 51:409–20.

12. Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013; 73:5609–12.

13. Han D, Li J, Wang H, Su X, Hou J, Gu Y, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017; 66:1151–64.

14. Li H, Yang J, Wei X, Song C, Dong D, Huang Y, et al. Circ-FUT10 reduces proliferation and facilitates differentiation of myoblasts by sponging miR-133a. J Cell Physiol. 2018; 233:4643–51.

15. Yao JT, Zhao SH, Liu QP, Lv MQ, Zhou DX, Liao ZJ, et al. Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract. 2017; 213:453–6.

16. Fu L, Yao T, Chen Q, Mo X, Hu Y, Guo J. Screening differential circular RNA expression profiles reveals hsa_circ_0004018 is associated with hepatocellular carcinoma. Oncotarget. 2017; 8:58405–16.

17. Li P, Chen H, Chen S, Mo X, Li T, Xiao B, et al. Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br J Cancer. 2017; 116:626–33.

18. Li J, Yang J, Zhou P, Le Y, Zhou C, Wang S, et al. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am J Cancer Res. 2015; 5:472–80.
19. Liang HF, Zhang XZ, Liu BG, Jia GT, Li WL. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res. 2017; 7:1566–76.
20. Tang YY, Zhao P, Zou TN, Duan JJ, Zhi R, Yang SY, et al. Circular RNA hsa_circ_0001982 promotes breast cancer cell carcinogenesis through decreasing miR-143. DNA Cell Biol. 2017; 36:901–8.

21. Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep. 2016; 6:30919.

22. Zou CD, Zhao WM, Wang XN, Li Q, Huang H, Cheng WP, et al. MicroRNA-107: a novel promoter of tumor progression that targets the CPEB3/EGFR axis in human hepatocellular carcinoma. Oncotarget. 2016; 7:266–78.

23. Wang S, Ma G, Zhu H, Lv C, Chu H, Tong N, et al. miR-107 regulates tumor progression by targeting NF1 in gastric cancer. Sci Rep. 2016; 6:36531.

24. Milella M, Falcone I, Conciatori F, Cesta Incani U, Del Curatolo A, Inzerilli N, et al. PTEN: multiple functions in human malignant tumors. Front Oncol. 2015; 5:24.
25. Yu G, Chen X, Chen S, Ye W, Hou K, Liang M. MiR-19a, miR-122 and miR-223 are differentially regulated by hepatitis B virus X protein and involve in cell proliferation in hepatoma cells. J Transl Med. 2016; 14:122.

26. Wei H, Cui R, Bahr J, Zanesi N, Luo Z, Meng W, et al. miR-130a deregulates PTEN and stimulates tumor growth. Cancer Res. 2017; 77:6168–78.
27. Wang S, Yuan L. Predictive biomarkers for targeted and cytotoxic agents in gastric cancer for personalized medicine. Biosci Trends. 2016; 10:171–80.

28. Chen J, Cui L, Yuan J, Zhang Y, Sang H. Circular RNA WDR77 target FGF-2 to regulate vascular smooth muscle cells proliferation and migration by sponging miR-124. Biochem Biophys Res Commun. 2017; 494:126–32.

Fig. 1.
Circular RNA ZFR (circ-ZFR) was significantly downregulated in gastric cancer (GC) tissues and cells. (A, B) CircZFR was low-expressed in GC tumor tissues compared with adjacent tissues confirmed by microarray analysis and shown in Volcano plot and heat map. (C) Circ-ZFR was significantly downregulated in tumor tissues compared with adjacent tissues. (D) The expression of circ-ZFR was considerably lower in human GC cell lines (AGS, AZ521, and HGC-27) than in gastric epithelial cell line GES-1 determined by real-time reverse transcription polymerase chain reaction. Among three GC cell lines, AGS cell line presented the most significant difference of circ-ZFR expression in comparison with GES-1 cell line. (E) The presence of circ-ZFR in AGS cells showed that there was no less expression afterthe RNase was processed. GAPDH, glyceraldehyde 3-phosphate dehydrogenase. *p < 0.05, **p < 0.01, compared with adjacent tissues or GES-1 or normal control (NC).
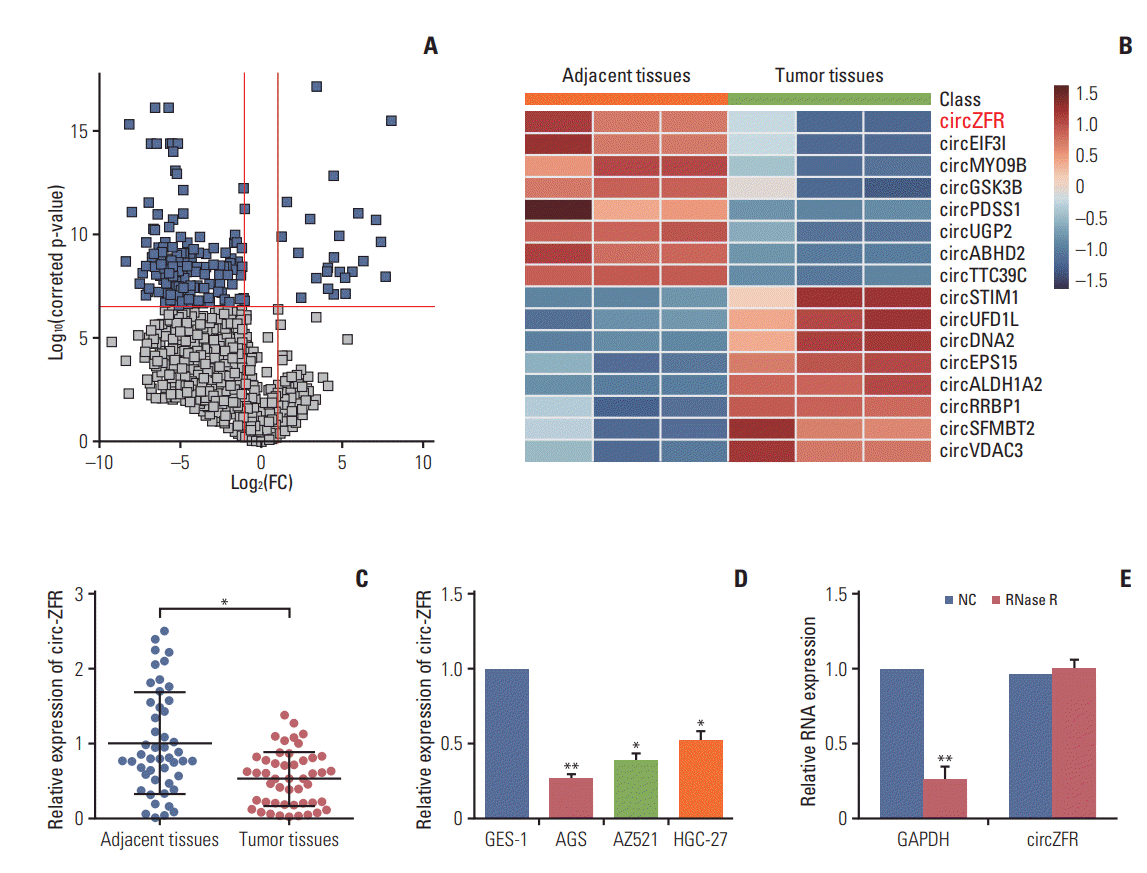
Fig. 2.
Overexpression of circular RNA ZFR (circ-ZFR) impeded cell reproduction and cell cycle and promoted apoptosis in gastric cancer (GC). (A) Circ-ZFR expression was remarkably upregulated after transfection with pcDNA3.1-circZFR examined by real-time reverse transcription polymerase chain reaction. (B) Overexpression of circ-ZFR observably suppressed GC cell proliferation, as confirmed in cell counting kit 8 assay. (C) The proportion of cells arrested in G0/G1 phase was significantly higher in pcDNA3.1-circZFR group than in normal control (NC) group, while that of arrested cells in S phase relatively reduced compared with NC group analyzed by flow cytometry assay. (D) The apoptosis rate of GC cells significantly increased after transfection with pcDNA3.1-circZFR detected by flow cytometry assay. PI, propidium iodide. *p < 0.05, **p < 0.01, compared with NC group.
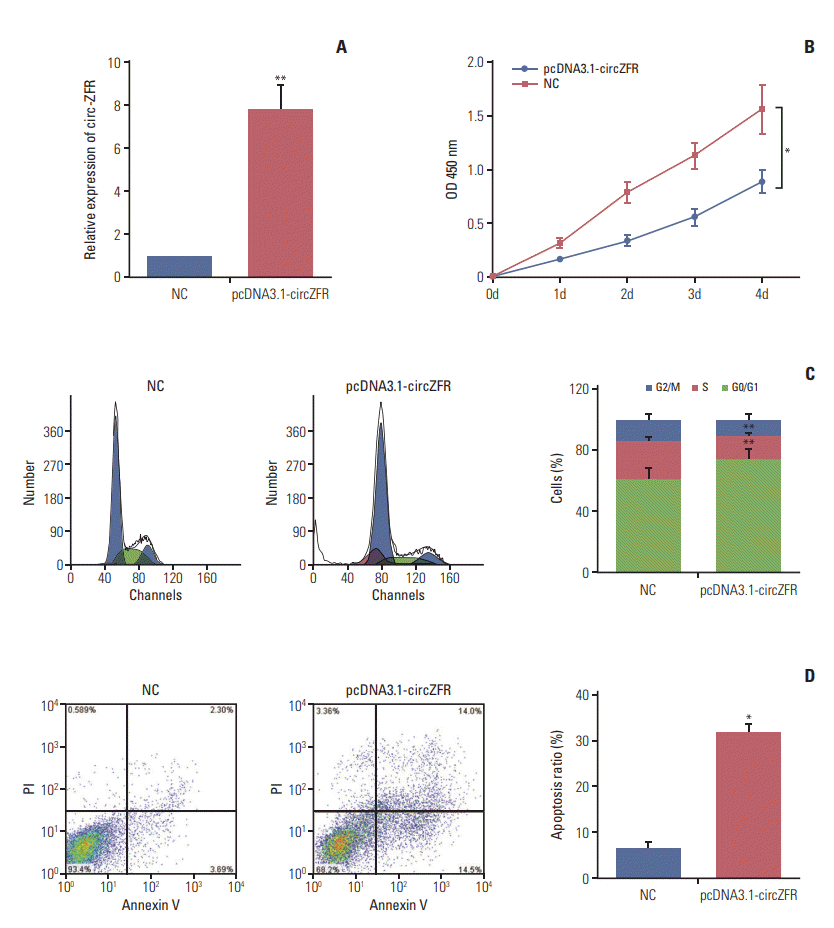
Fig. 3.
Prediction of circRNA-miRNA-mRNA associations in gastric cancer. (A) The binding sites of miR-107/miR-130a on
3′-untranslated regions (3′-UTR) of circular RNA ZFR (circ-ZFR) was confirmed by starbase. (B) The schematic model showing
the putative binding sites for miRNAs and 3′-untranslated region of circ-ZFR. (C) The pathways co-affected by miR-107
and miR-130a were displayed by DIANA TOOLS-mirPath v.3. (D) The crossed targeting gene regulated by both p53 pathway
and miR-130a/miR-107 via Veen diagram. (E) The binding sites of miR-107/miR-130a on 3′-UTR of PTEN was validated by
TargetScan. (F) A biomathematically predicted target network between circ-ZFR, miR-130a/miR-107, and PTEN.
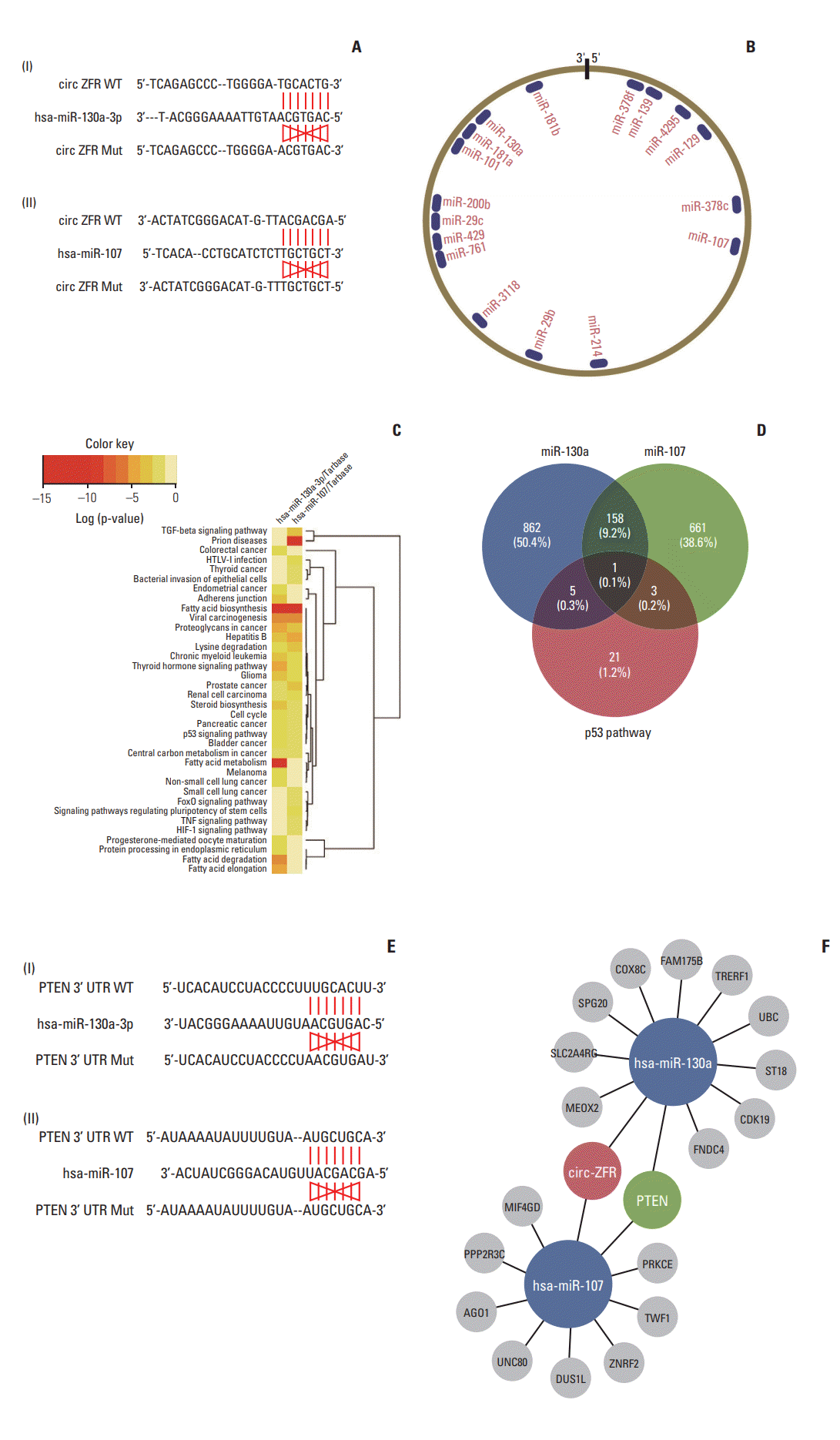
Fig. 4.
Confirmation of targeted relationships between miR-107/miR-130a and PTEN or circular RNA ZFR (circ-ZFR). (A) MiR-107 mimics and miR-130a mimics led to overexpression of miR-107 and miR-130a, respectively detected by real-time reverse transcription polymerase chain reaction. (B) The expression of circ-ZFR was no significant difference both in miR107‒mimics and miR-130a–mimics group. (C, D) Overexpression of miR-107 and miR-130a both significantly repressed the luciferase activity of the luciferase reporter containing ZFR 3′-untranslated regions (3′-UTR)‒WT but not the reporter containing ZFR 3′-UTR‒Mut confirmed by dual luciferase reporter assay. (E, F) There existed some interactions between ZFR and miR-107/miR-130a verified by RNA immunoprecipitation assay.
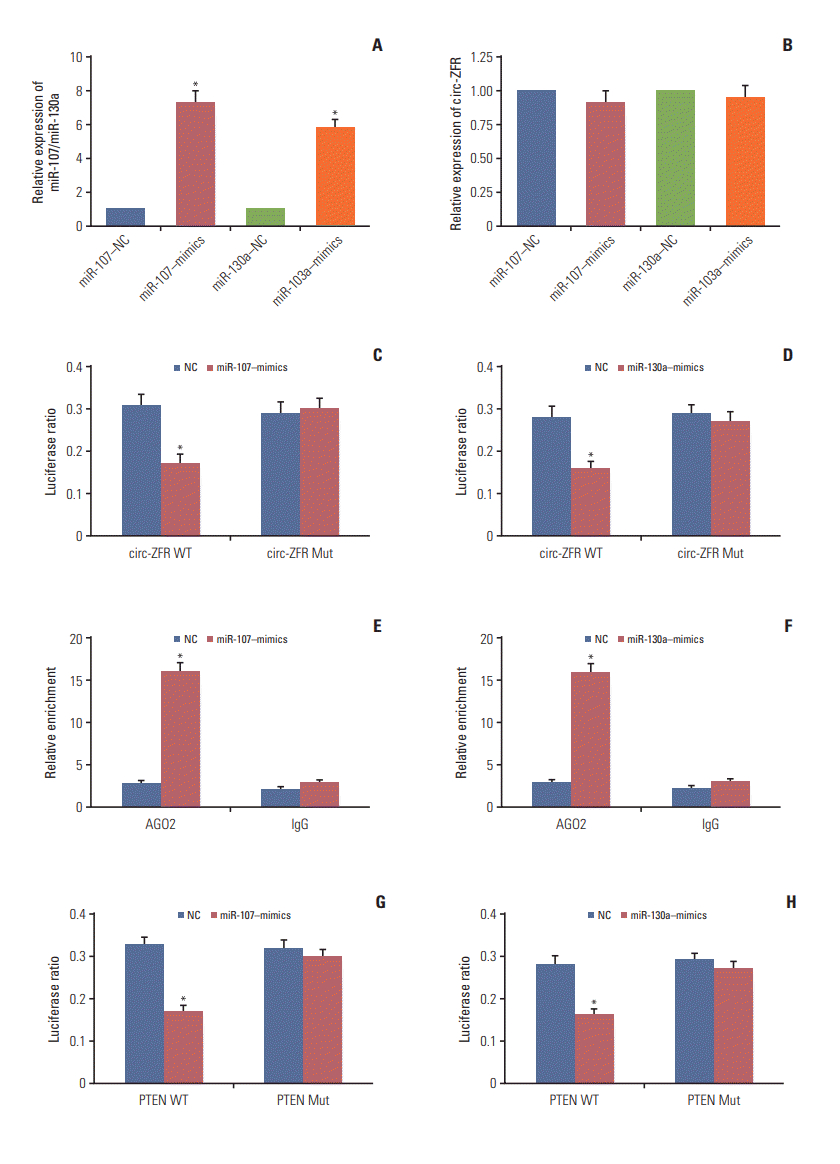
Fig. 5.
Correlations on expression between miR-107/miR-130a and PTEN/circ-ZFR. (A, B) MiR-107 and miR-130a were highly expressed whereas PTEN was lowly expressed in tumor tissues compared with adjacent tissues examined by realtime reverse transcription polymerase chain reaction. (C, D) The expressions of miR-107 and miR-130a were both negatively correlated with ZFR expression determined by Pearson correlation analysis. (E, F) The expressions of miR-107 and miR-130a were both negatively correlated with PTEN expression determined by Pearson correlation analysis. *p < 0.05, compared with adjacent tissues. GC, gastric cancer.
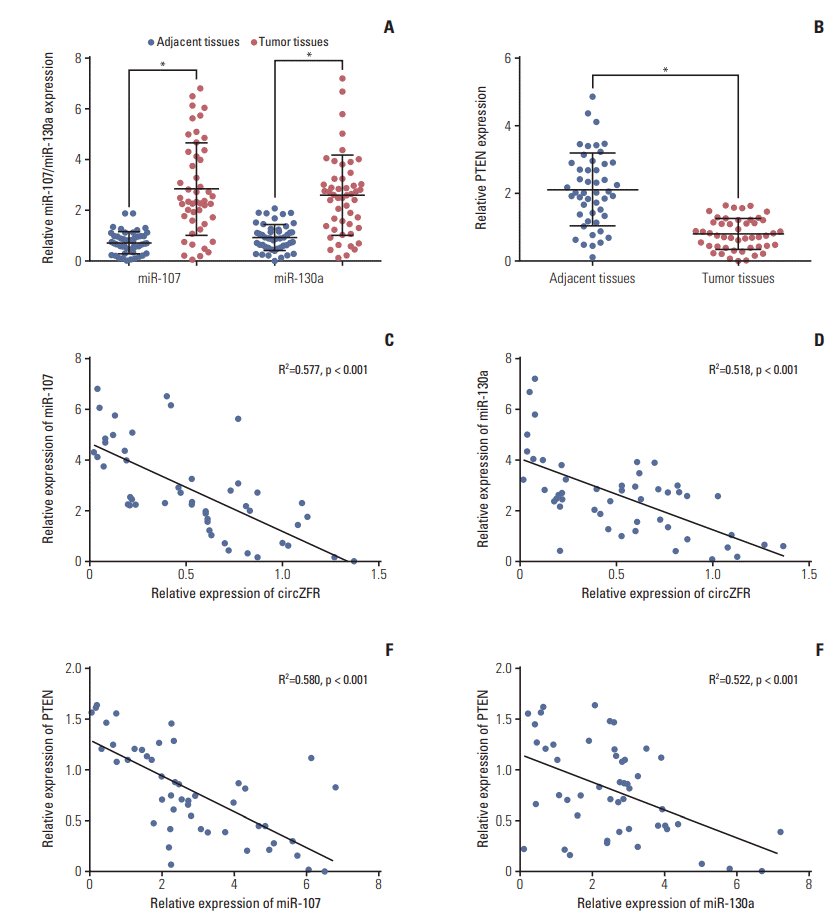
Fig. 6.
Circ-ZFR suppressed gastric cancer cell reproduction by sponging miR-107/miR-130a. (A, C) MiR-107/miR-130a mimics significantly increased the expression of miR-107/miR-130a, while miR-107/miR-130a inhibitor remarkably decreased miR-107/miR-130a expressions examined by real-time reverse transcription polymerase chain reaction. (B, D) Overexpression of miR-107 and miR-130a both promoted cell proliferation whereas downregulation of miR-107 and miR130a expressions restrained propagation. No significant difference of cell proliferation was found between miR-107/miR130a+pcDNA3.1-ZFR group and normal control (NC) group. However, the proliferation ability of the cells in miR-107/miR-130a+pcDNA3.1-ZFR group was significantly stronger than that in pcDNA3.1-ZFR group, while weaker compared with miR-107/miR-130a–mimics group. *p < 0.05, **p < 0.01, compared with NC group; #p < 0.05, compared with pcDNA3.1-ZFR group; †p < 0.05, compared with miR-107/miR-130a–mimics group.
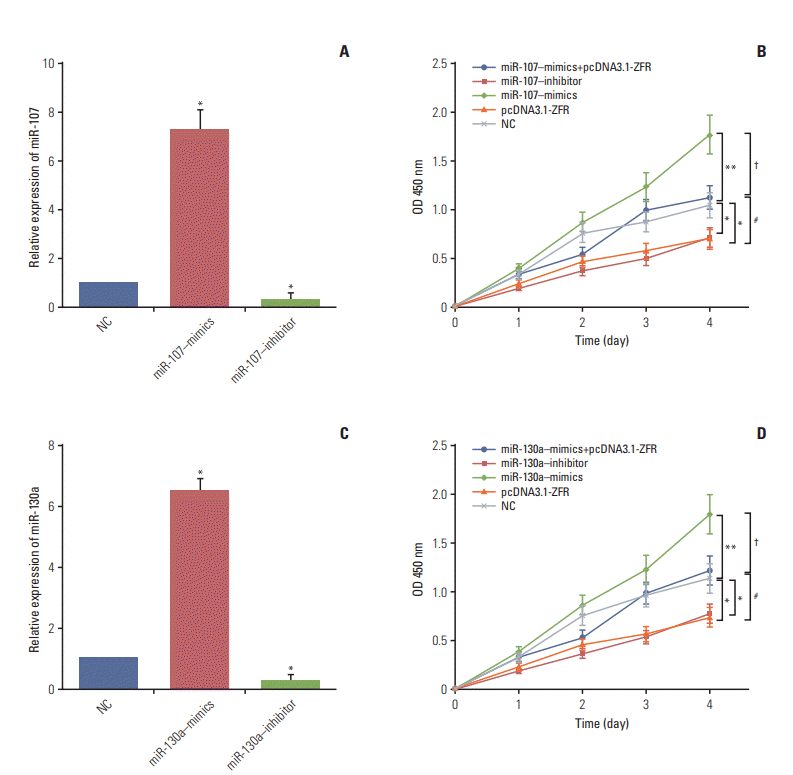
Fig. 7.
Circular RNA ZFR (circ-ZFR) induced cell cycle arrest by sponging miR-107/miR-130a in gastric cancer. (A, B) The
proportion of the cells arrested in G1 phase in pcDNA3.1-ZFR group, miR-107 inhibitor group and miR-130a inhibitor group was considerably larger than in normal control (NC) group. Conversely, the percentage of arrested cell in G1 phase in miR107–mimics group and miR-130a–mimics group was relatively smaller compared with NC group. Nonetheless, the percentage of arrested cell in G1 phase in miR-107/miR-130a+pcDNA3.1-ZFR group was significantly smaller than that in pcDNA3.1-ZFR group, while larger than that in miR-107/miR-130a–mimics group. There was no conspicuous distinction between miR-107/miR-130a+pcDNA3.1-ZFR group and NC group. *p < 0.05, compared with NC group; #p < 0.05, compared with pcDNA3.1-ZFR group; †p < 0.05, compared with miR-107/miR-130a–mimics group.
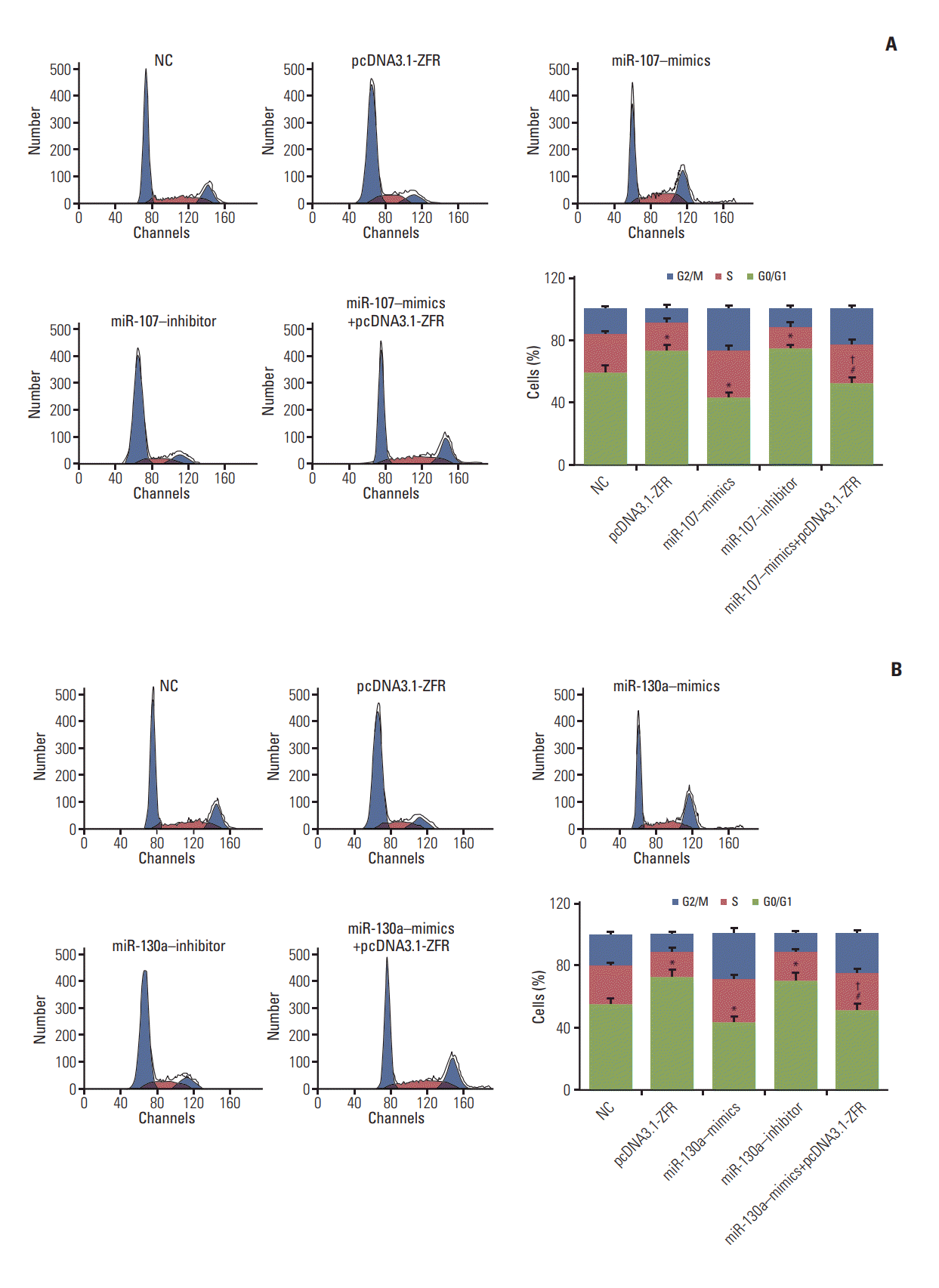
Fig. 8.
Circular RNA ZFR (circ-ZFR) facilitated gastric cancer cell apoptosis by sponging miR-107/miR-130a (A, B), The apoptosis of cells was significantly enhanced after transfected with pcDNA3.1-ZFR and miR-107/miR-130a inhibitor, while that of the cells transfected with miR-107/miR-130a–mimics was remarkably inhibited. No significant difference of apoptosis rate was detected between miR-107/miR-130a+pcDNA3.1-ZFR group and normal control (NC) group. However, the cell apoptosis rate in miR-107/miR-130a+pcDNA3.1-ZFR group was considerably lower than that in pcDNA3.1-ZFR group, while significantly higher compared with miR-107/miR-130a‒mimics group. *p < 0.05, ** p < 0.01, compared with NC group; #p < 0.05, compared with pcDNA3.1-ZFR group; †p < 0.05, compared with miR-107/miR-130a–mimics group.
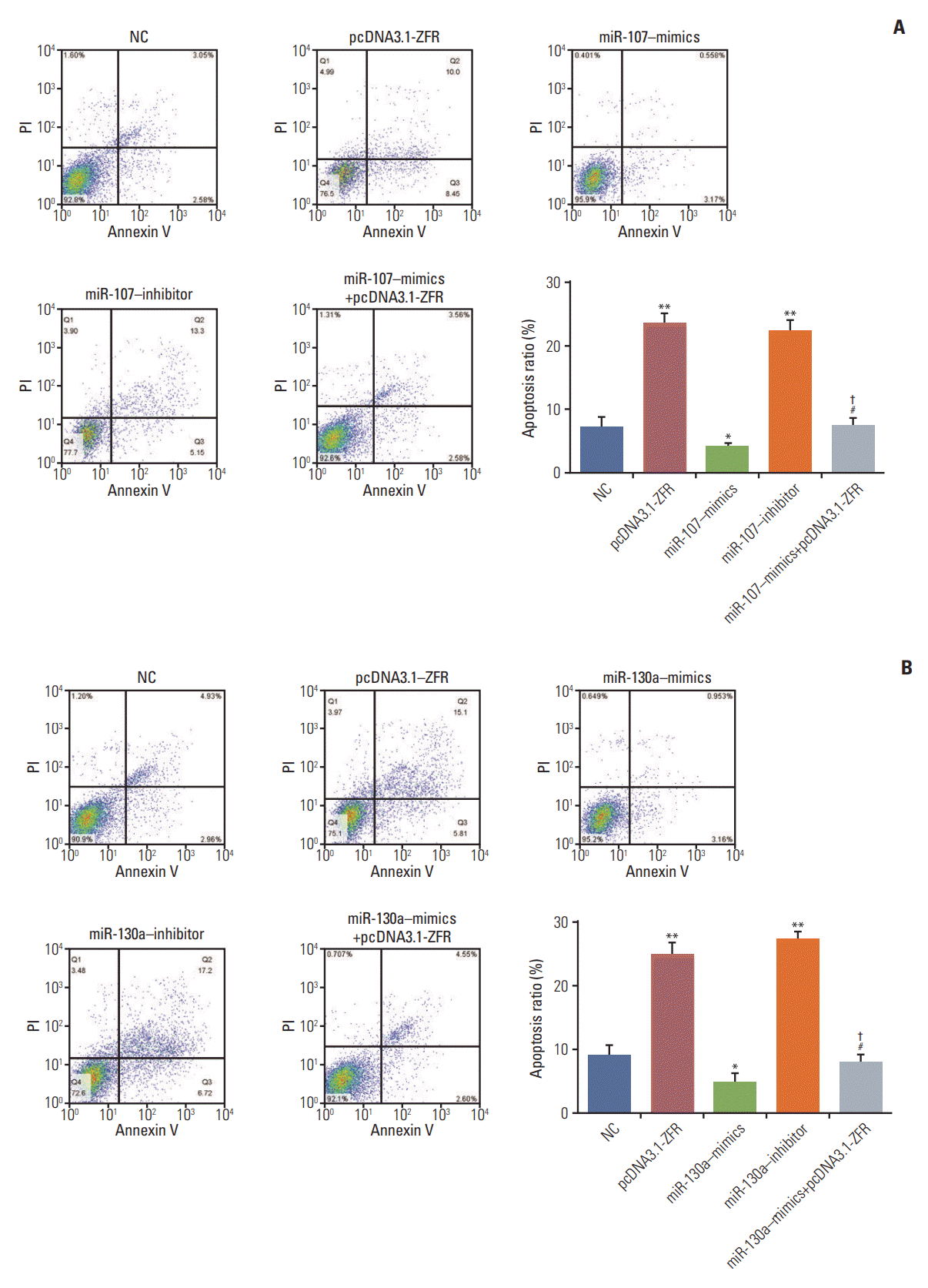
Fig. 9.
miR-107/miR-130a promoted gastric cancer cell reproduction through targeting PTEN. (A, B) The expression of PTEN was remarkably repressed after transfection with miR-107‒mimics/miR-130a‒mimics, while it was overexpressed followed transfection with pcDNA3.1-ZFR and miR-107 inhibitor/miR-130a inhibitor detected with western blot. (C) The expression of pcDNA3.1-PTEN in gastric cancer cells was dramatically upregulated after transfection with pcDNA3.1-PTEN examined by real-time reverse transcription polymerase chain reaction. (D, E) The proliferation ability of the cells in miR-107/miR130a‒mimics+pcDNA3.1-PTEN group was significantly stronger than that in pcDNA3.1-PTEN group, while weaker compared with miR-107/miR-130a‒mimics group. No significant difference of cell proliferation was observed between miR-107/miR-130a‒mimics+pcDNA3.1-PTEN group and normal control (NC) group. *p < 0.05, compared with NC group; #p < 0.05, compared with pcDNA3.1-PTEN group; † p < 0.05, compared with miR-107/miR-130a‒mimics group.
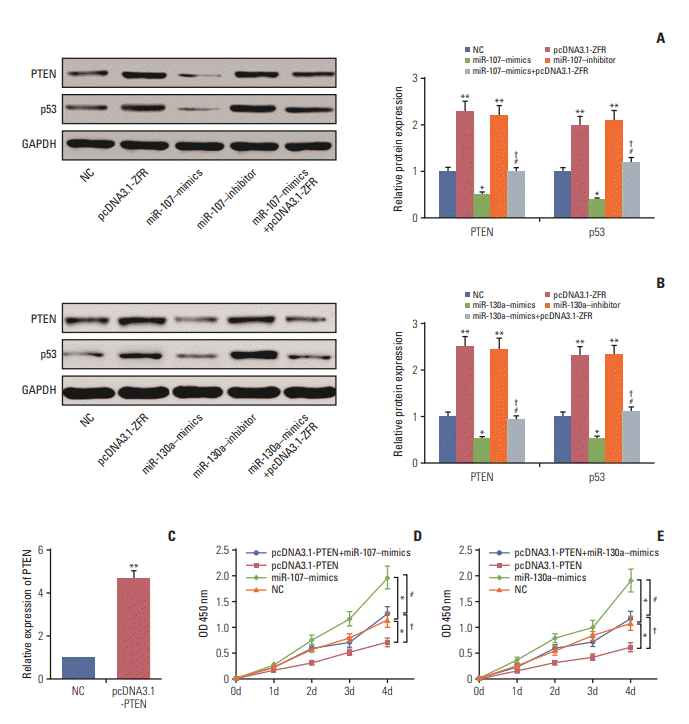
Fig. 10.
miR-107/miR-130a influenced gastric cancer cell cycle through targeting PTEN. (A, B) Flow cytometry assay displayed that the proportion of the cells arrested in G1 phase in miR-107/miR-130a–mimics group dramatically reduced compared with normal control (NC) group, while that in S phase increased and there was no significant difference in G2 phase. On the contrary, the percentage of arrested cell in G1 phase in pcDNA3.1-PTEN group conspicuously increased in comparison with NC group whereas that in S phase relatively decreased. Nonetheless, the percentage of arrested cell in G1 phase in miR-107/miR-130a+pcDNA3.1-PTEN group was significantly smaller than that in pcDNA3.1-PTEN group, while larger than that in miR-107/miR-130a–mimics group. There was no significant distinction between miR-107/miR-130a+pcDNA3.1-PTEN group and NC group. *p < 0.05, compared with NC group; #p < 0.05, compared with pcDNA3.1-PTEN group; †p < 0.05, compared with miR-107/miR-130a–mimics group.
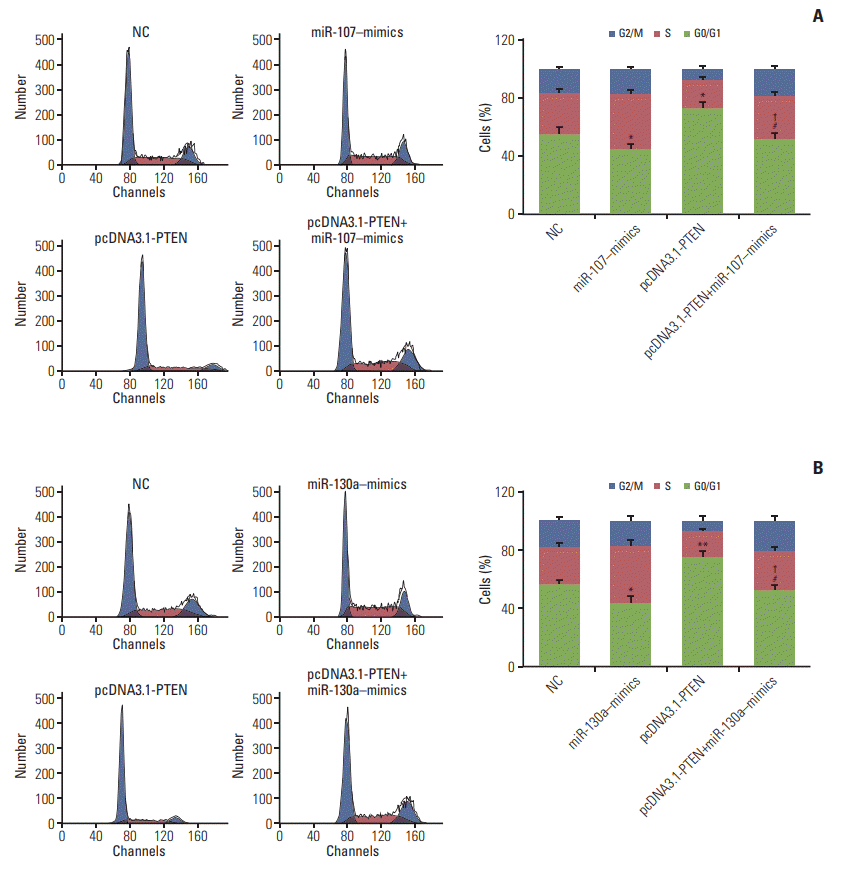
Fig. 11.
miR-107/miR-130a impeded cell apoptosis in gastric cancer through targeting PTEN. (A, B) The apoptotic rate of the cells transfected with miR-107/miR-130a–mimics considerably fell whereas that of the cells transfected with pcDNA3.1-PTEN drastically rose. No significant difference of apoptosis ratio was found between miR-107/miR-130a+pcDNA3.1-PTEN group and normal control (NC) group. Nevertheless, the cell apoptosis rate in miR-107/miR-130a+pcDNA3.1-PTEN group was considerably lower than that in pcDNA3.1-PTEN group, while significantly higher compared with miR-107/miR-130a–mimics group. *p < 0.05, compared with NC group; #p < 0.05, compared with pcDNA3.1-PTEN group; †p < 0.05, compared with miR-107/miR-130a–mimics group.
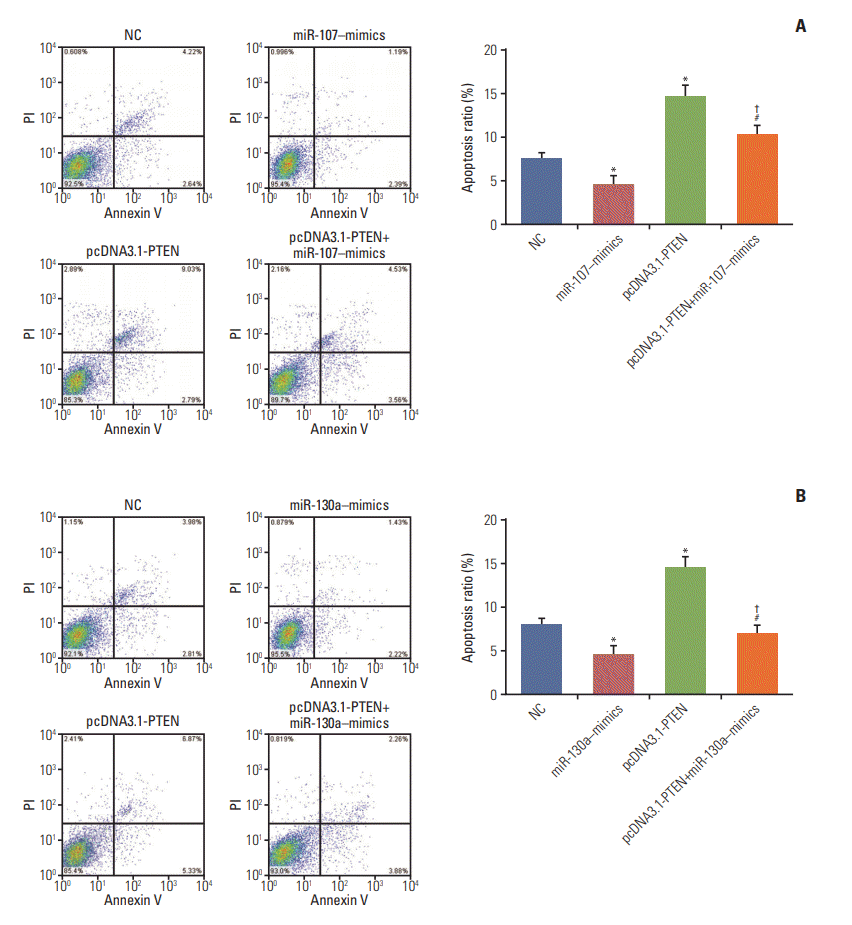
Fig. 12.
Circular RNA ZFR curbed gastric cancer (GC) tumor growth and affected p53 protein expression in vivo (A) The tumor volume of the mice in pcDNA3.1-ZFR group was significantly grew more slowly than that in normal control (NC) group. (B, C) The tumor weight of mice in pcDNA3.1-ZFR group was significantly lighter compared with NC group. (D, E) The expression level of miR-107 (D) and miR-130a (E) was decreased after transfected with pcDNA3.1-ZFR. (F) The expression level of p53 protein dramatically increased after transfected with pcDNA3.1-ZFR confirmed by immunohistochemistry (IHC) assay. (G) The expression level of PTEN increased after transfected with pcDNA3.1-ZFR confirmed by IHC assay. *p < 0.05, **p < 0.01, compared with NC group.
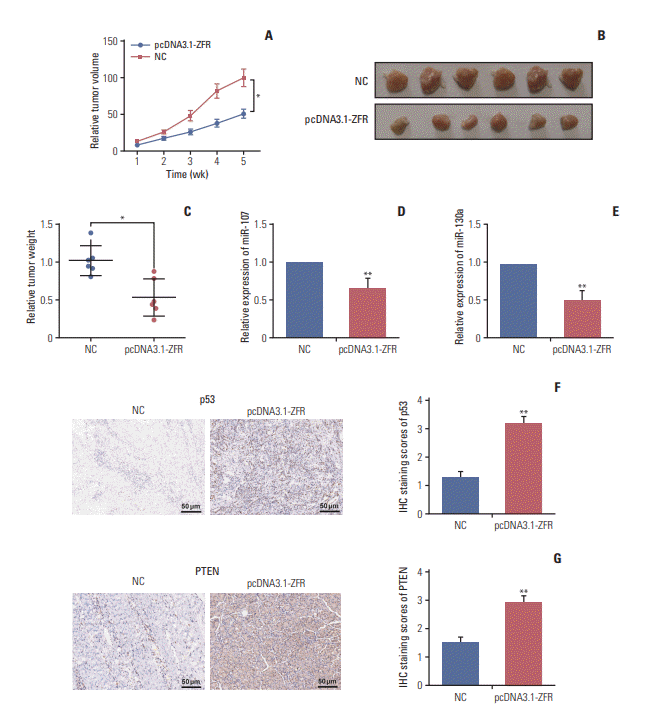
Fig. 13.
The mechanism of circular RNA ZFR (circ-ZFR)–miR-107/miR-130a–PTEN axis. Circ-ZFR was identified as a sponge of miR-130a/miR-107 and indirectly modulated their target PTEN expression. GC, gastric cancer.
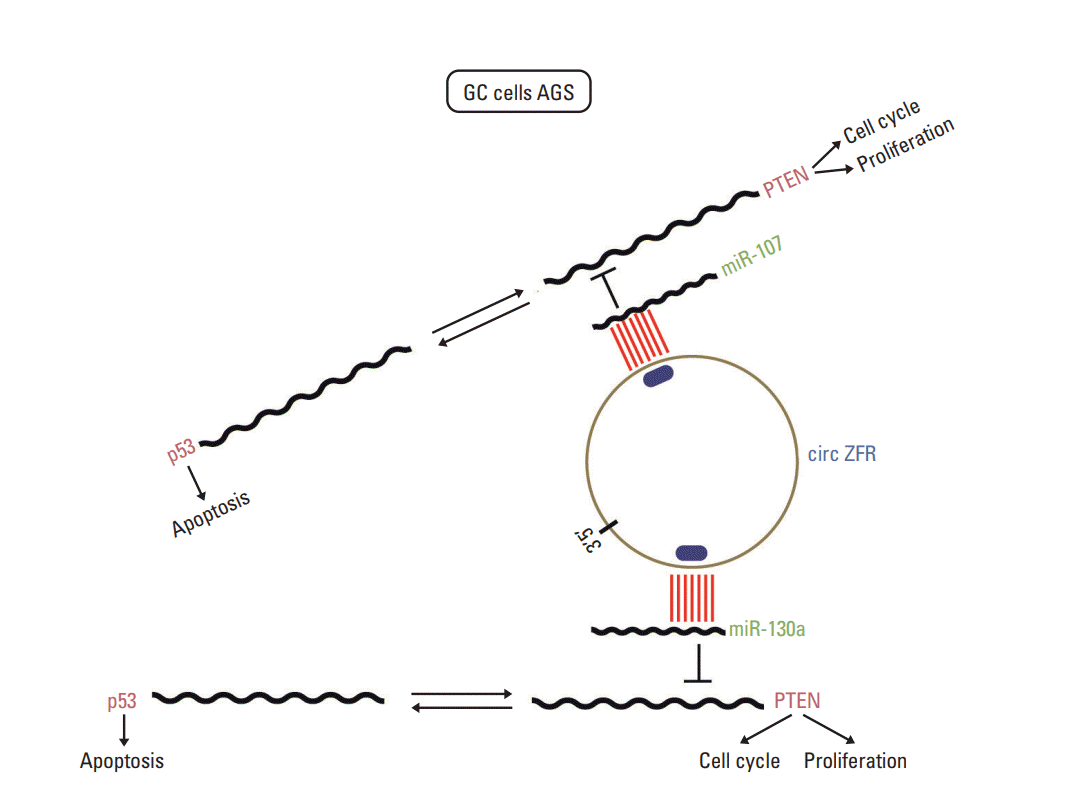
Table 1.
Polymerase chain reaction primer sequences



