1. Spano JP, Lagorce C, Atlan D, Milano G, Domont J, Benamouzig R, et al. Impact of EGFR expression on colorectal cancer patient prognosis and survival. Ann Oncol. 2005; 16:102–8.

2. Saijo N. Present status and problems on molecular targeted therapy of cancer. Cancer Res Treat. 2012; 44:1–10.

3. Kim D, Hong YS, Kim JE, Kim KP, Lee JL, Chun SM, et al. Use of a High-throughput genotyping platform (OncoMap) for RAS mutational analysis to predict cetuximab efficacy in patients with metastatic colorectal cancer. Cancer Res Treat. 2017; 49:37–43.
4. Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008; 359:1757–65.
5. Schneider MR, Wolf E. The epidermal growth factor receptor ligands at a glance. J Cell Physiol. 2009; 218:460–6.

6. Khambata-Ford S, Garrett CR, Meropol NJ, Basik M, Harbison CT, Wu S, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol. 2007; 25:3230–7.

7. Hobor S, Van Emburgh BO, Crowley E, Misale S, Di Nicolantonio F, Bardelli A. TGFα and amphiregulin paracrine network promotes resistance to EGFR blockade in colorectal cancer cells. Clin Cancer Res. 2014; 20:6429–38.

8. Khelwatty S, Essapen S, Bagwan I, Green M, Seddon A, Modjtahedi H. The impact of co-expression of wild-type EGFR and its ligands determined by immunohistochemistry for response to treatment with cetuximab in patients with metastatic colorectal cancer. Oncotarget. 2017; 8:7666–77.

9. Ohchi T, Akagi Y, Kinugasa T, Kakuma T, Kawahara A, Sasatomi T, et al. Amphiregulin is a prognostic factor in colorectal cancer. Anticancer Res. 2012; 32:2315–21.
10. Jacobs B, De Roock W, Piessevaux H, Van Oirbeek R, Biesmans B, De Schutter J, et al. Amphiregulin and epiregulin mRNA expression in primary tumors predicts outcome in metastatic colorectal cancer treated with cetuximab. J Clin Oncol. 2009; 27:5068–74.

11. Jonker DJ, Karapetis CS, Harbison C, O'Callaghan CJ, Tu D, Simes RJ, et al. Epiregulin gene expression as a biomarker of benefit from cetuximab in the treatment of advanced colorectal cancer. Br J Cancer. 2014; 110:648–55.

12. Fujiwara S, Hung M, Yamamoto-Ibusuk CM, Yamamoto Y, Yamamoto S, Tomiguchi M, et al. The localization of HER4 intracellular domain and expression of its alternately-spliced isoforms have prognostic significance in ER+ HER2- breast cancer. Oncotarget. 2014; 5:3919–30.

13. Yun S, Koh J, Nam SK, Park JO, Lee SM, Lee K, et al. Clinical significance of overexpression of NRG1 and its receptors, HER3 and HER4, in gastric cancer patients. Gastric Cancer. 2018; 21:225–36.

14. Wang J, Yin J, Yang Q, Ding F, Chen X, Li B, et al. Human epidermal growth factor receptor 4 (HER4) is a favorable prognostic marker of breast cancer: a systematic review and meta-analysis. Oncotarget. 2016; 7:76693–703.

15. Lee KS, Nam SK, Koh J, Kim DW, Kang SB, Choe G, et al. Stromal expression of microRNA-21 in advanced colorectal cancer patients with distant metastases. J Pathol Transl Med. 2016; 50:270–7.

16. Seo AN, Kwak Y, Kim WH, Kim DW, Kang SB, Choe G, et al. HER3 protein expression in relation to HER2 positivity in patients with primary colorectal cancer: clinical relevance and prognostic value. Virchows Arch. 2015; 466:645–54.

17. Atkins D, Reiffen KA, Tegtmeier CL, Winther H, Bonato MS, Storkel S. Immunohistochemical detection of EGFR in paraffin-embedded tumor tissues: variation in staining intensity due to choice of fixative and storage time of tissue sections. J Histochem Cytochem. 2004; 52:893–901.
18. Chakraborty S, Li L, Puliyappadamba VT, Guo G, Hatanpaa KJ, Mickey B, et al. Constitutive and ligand-induced EGFR signalling triggers distinct and mutually exclusive downstream signalling networks. Nat Commun. 2014; 5:5811.

19. Guo G, Gong K, Wohlfeld B, Hatanpaa KJ, Zhao D, Habib AA. Ligand-independent EGFR signaling. Cancer Res. 2015; 75:3436–41.

20. Schrevel M, Osse EM, Prins FA, Trimbos JB, Fleuren GJ, Gorter A, et al. Autocrine expression of the epidermal growth factor receptor ligand heparin-binding EGF-like growth factor in cervical cancer. Int J Oncol. 2017; 50:1947–54.

21. Iwamoto R, Takagi M, Akatsuka J, Ono K, Kishi Y, Mekada E. Characterization of a novel anti-human HB-EGF monoclonal antibody applicable for paraffin-embedded tissues and diagnosis of HB-EGF-related cancers. Monoclon Antib Immunodiagn Immunother. 2016; 35:73–82.

22. Zhou ZN, Sharma VP, Beaty BT, Roh-Johnson M, Peterson EA, Van Rooijen N, et al. Autocrine HBEGF expression promotes breast cancer intravasation, metastasis and macrophage-independent invasion in vivo. Oncogene. 2014; 33:3784–93.

23. Byeon SJ, Lee HS, Kim MA, Lee BL, Kim WH. Expression of the ERBB family of ligands and receptors in gastric cancer. Pathobiology. 2017; 84:210–7.

24. Kawasaki H, Saotome T, Usui T, Ohama T, Sato K. Regulation of intestinal myofibroblasts by KRas-mutated colorectal cancer cells through heparin-binding epidermal growth factor-like growth factor. Oncol Rep. 2017; 37:3128–36.

25. Murata T, Mizushima H, Chinen I, Moribe H, Yagi S, Hoffman RM, et al. HB-EGF and PDGF mediate reciprocal interactions of carcinoma cells with cancer-associated fibroblasts to support progression of uterine cervical cancers. Cancer Res. 2011; 71:6633–42.

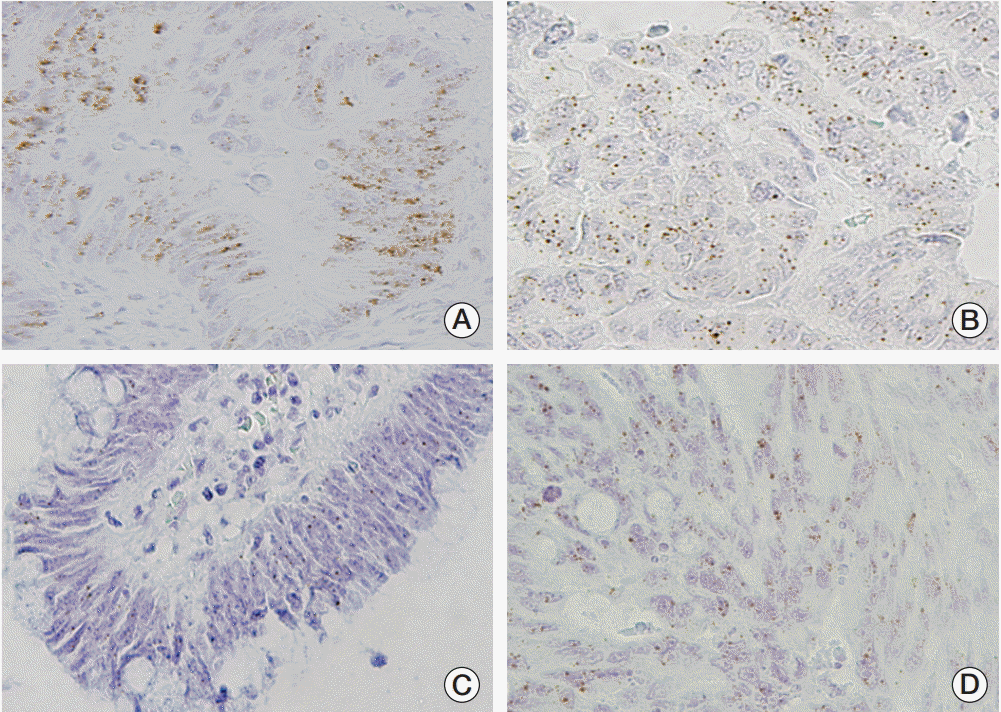
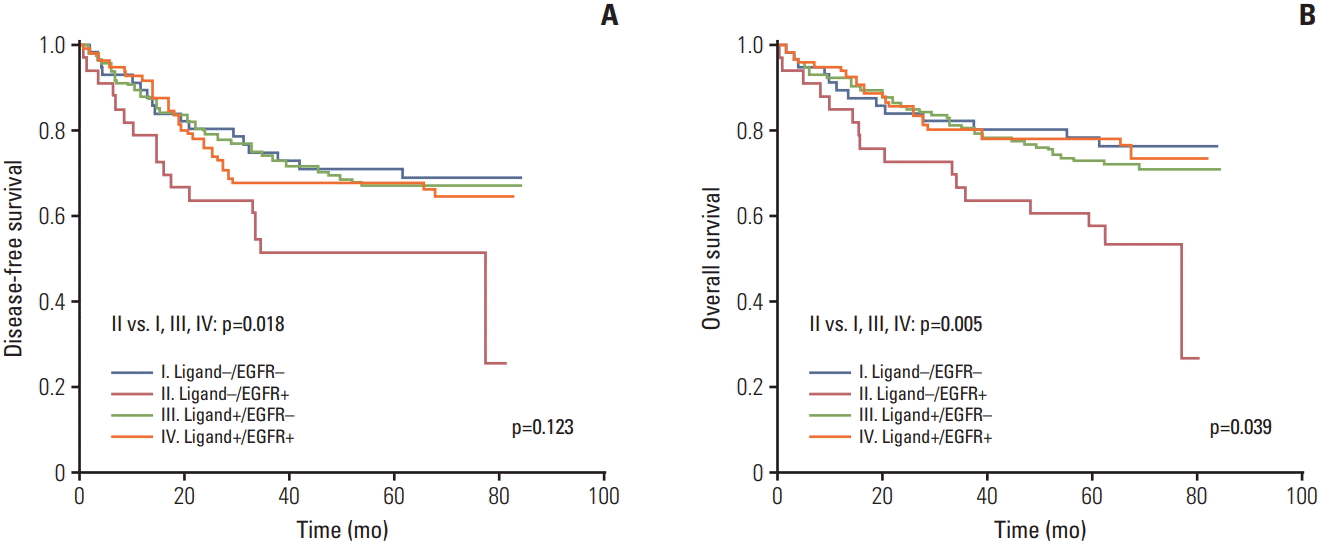
26. Yoshida M, Shimura T, Sato M, Ebi M, Nakazawa T, Takeyama H, et al. A novel predictive strategy by immunohistochemical analysis of four EGFR ligands in metastatic colorectal cancer treated with anti-EGFR antibodies. J Cancer Res Clin Oncol. 2013; 139:367–78.

27. Ito M, Yoshida K, Kyo E, Ayhan A, Nakayama H, Yasui W, et al. Expression of several growth factors and their receptor genes in human colon carcinomas. Virchows Arch B Cell Pathol Incl Mol Pathol. 1990; 59:173–8.

28. Rajagopal S, Huang S, Moskal TL, Lee BN, el-Naggar AK, Chakrabarty S. Epidermal growth factor expression in human colon and colon carcinomas: anti-sense epidermal growth factor receptor RNA down-regulates the proliferation of human colon cancer cells. Int J Cancer. 1995; 62:661–7.

29. Mota JM, Collier KA, Barros Costa RL, Taxter T, Kalyan A, Leite CA, et al. A comprehensive review of heregulins, HER3, and HER4 as potential therapeutic targets in cancer. Oncotarget. 2017; 8:89284–306.





 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download