Introduction
Materials and Methods
1. Samples and DNA/RNA extraction
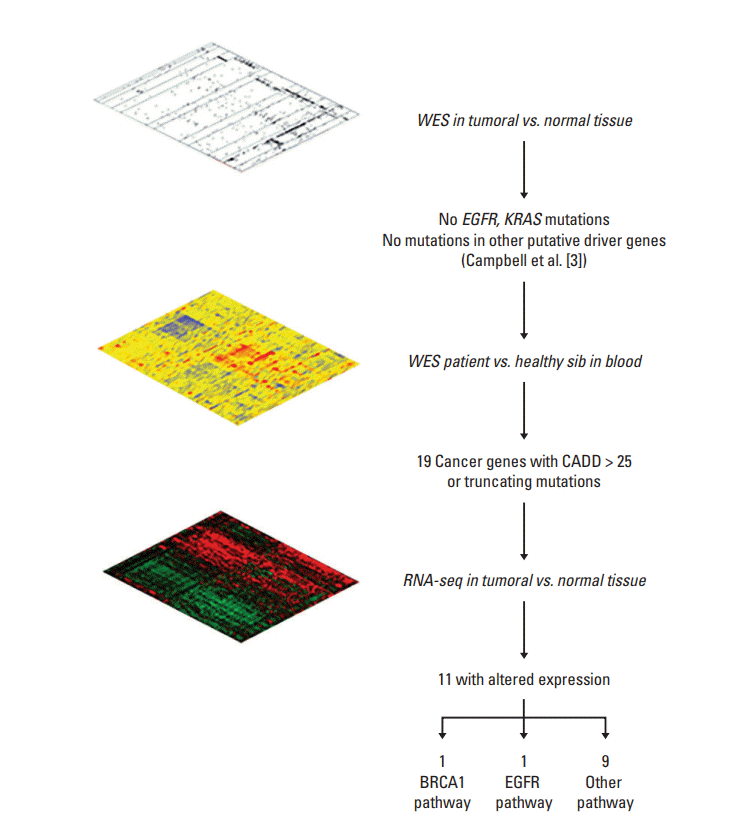
2. WES and data analysis
3. RNA-seq and data analysis
Results
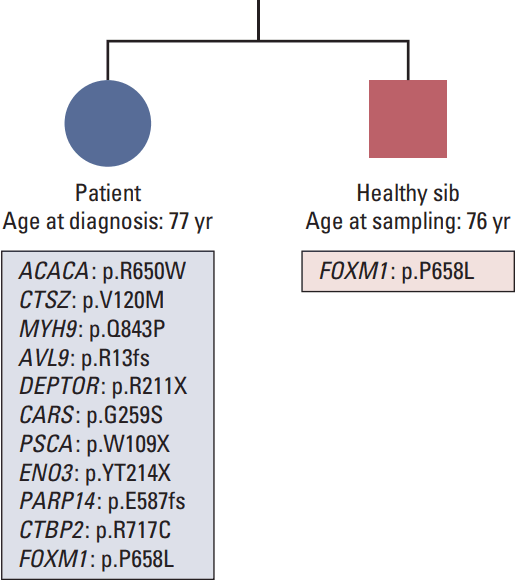
1. Patient description
2. “Omic” characterization
Table 1.
| Group | Gene symbol | Amino acid change (CADD value) | Fold-change T/N | Gene function related to cancer development |
|---|---|---|---|---|
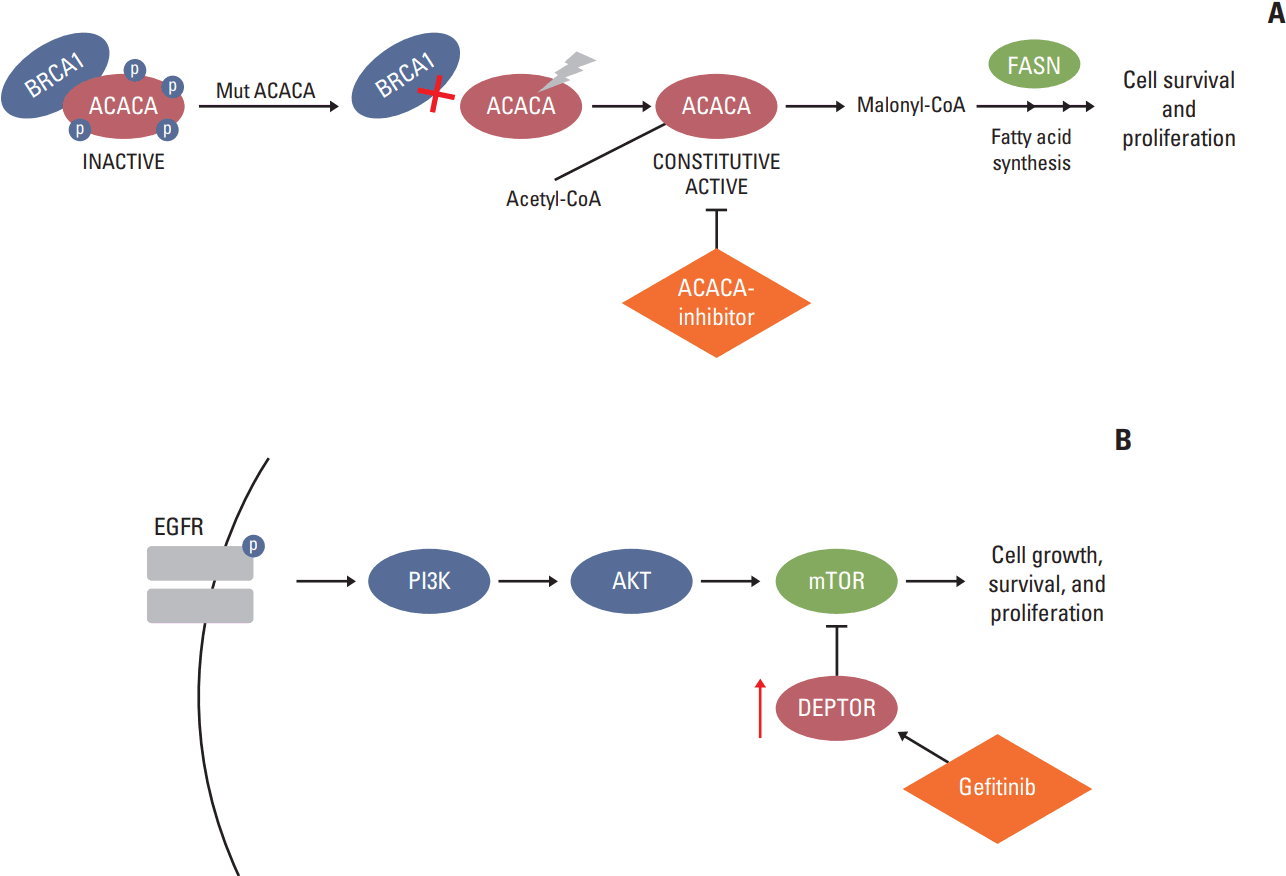
| Group 1: Underexpressed in tumoral tissue (fold-change ≤ 0.66) | ACACA | p.R650W (33) | –4.35 | Acetyl-CoA carboxylase alpha has a role in de novo lipogenesis catalyzing the carboxylation of acetyl-CoA to malonyl-CoA. The enzyme is under long term control at the transcriptional and translational levels. It is known to specifically interact with BRCA1 [11]. |
| MYH9 | p.Q843P (31) | –1.64 | Myosin heavy chain 9 is a myosin IIA heavy chain that interacts with β-actin to maintain cytoskeleton integrity. It is a EGFR-associating protein in NSCLC cell lines overexpressing EGFR [12]. | |
| CTSZ | p.V120M (26) | –1.59 | Cathepsin Z is a lysosomal cysteine proteinase. CTSZ is mostly expressed in immune cells, such as macrophages and monocytes, and involved in antigen processing and maturation of the major histocompatibility complex class II molecules [13]. This gene is expressed ubiquitously in cancer cell lines and primary tumors. | |
| Group 2: Underexpressed in both normal and tumoral tissue (fold-change ≤ 0.66) | AVL9 | p.R13fs (NA) | –3.13a) | AVL9 cell migration associated is among the 73 cancer driver candidate genes previously identified via dog-human comparison, deleted in colorectal cancer [14] and classified as putative tumor suppressors. AVL9-depletion inhibits the migration of A549 human adenocarcinomic alveolar basal epithelial cells [15]. |
| CARS | p.G259S (33)b) | –3.03a) | Cysteinyl-tRNA synthetase is one of several located near the imprinted gene domain on chromosome 11p15.5, an important tumor-suppressor gene region [16]. | |
| DEPTOR | p.R211X (46) | –1.96a) | DEP-domain containing mTOR interacting protein has growth suppression activity against pancreatic cancer cells, and its expression is gradually lost during pancreatic tumorigenesis [17]. Tumor progression mediated by EGFR ectopic expression was diminuished by transfection with DEPTOR in lung adenocarcinoma. Gefitinib, a specific EGFR inhibitor used for lung cancer therapy, stimulated DEPTOR accumulation [18]. | |
| Group 3: Overexpressed in tumoral tissue (fold-change ≥ +1.5) | PSCA | p.W109X (NA)b) | 391.96 | Prostate stem cell antigen encodes a glycosylphosphatidylinositol-anchored cell membrane glycoprotein that is upregulated in a large proportion of prostate cancers and other cancers. It is highly expressed in non-small cell lung cancer and may be functionally important for the disease: small interfering RNA-mediated knockdown of PSCA resulted in the inhibition of LC growth [19]. |
| ENO3 | p.YT214X (35) | 7.04 | Enolase 3 encodes one of the three enolase isoenzymes. It was found upregulated in liver cancer mouse model [20]. | |
| PARP14 | p.E587fs (NA) | 4.93 | Poly(ADP-ribose) polymerase family member 14 is an anti-apoptotic protein that may regulate aerobic glycolysis and promote survival of cancer cells [21]. Its increased expression has been reported in a variety of tumor types. | |
| CTBP2 | p.R717C (34) | 4.34 | C-terminal binding protein 2 is upregulated in a number of tumors, such as hepatocellular carcinoma, prostate cancer, and gliomas [22, 23]. | |
| FOXM1 | p.P658L (27)b) | 3.28 | Forkhead box M1 is a transcriptional activator involved in cell proliferation. Consistent with an important role of Foxm1 in cell cycle progression, increased expression of Foxm1 was found in many human tumors. Increased Foxm1 expression in human lung adenocarcinomas and squamous cell carcinomas was associated with increased proliferation of tumor cells [24]. |
CADD, Combined Annotation Dependent Depletion; T, tumoral lung tissue; N, non-tumoral lung tissue; ACACA, acetyl-CoA carboxylase alpha; EGFR, epidermal growth factor receptor; NSCLC, non-small cell lung cancers; mTOR, mammalian target of rapamycin; NA, not applicable; LC, lung cancer; MAF, minor allele frequency.




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download