Abstract
Since molecular biology studies began, researches in biological science have centered on proteins and genes at molecular level of a single cell. Cancer research has also focused on various functions of proteins and genes that distinguish cancer cells from normal cells. Accordingly, most contemporary anticancer drugs have been developed to target abnormal characteristics of cancer cells. Despite the great advances in the development of anticancer drugs, vast majority of patients with advanced cancer have shown grim prognosis and high rate of relapse. To resolve this problem, we must reevaluate our focuses in current cancer research. Cancer should be considered as a systemic disease because cancer cells undergo a complex interaction with various surrounding cells in cancer tissue and spread to whole body through metastasis under the control of the systemic modulation. Human body relies on the cooperative interaction between various tissues and organs, and each organ performs its specialized function through tissue-specific cell networks. Therefore, investigation of the tumor-specific cell networks can provide novel strategy to overcome the limitation of current cancer research. This review presents the limitations of the current cancer research, emphasizing the necessity of studying tissue-specific cell network which could be a new perspective on treating cancer disease, not cancer cells.
Go to : 
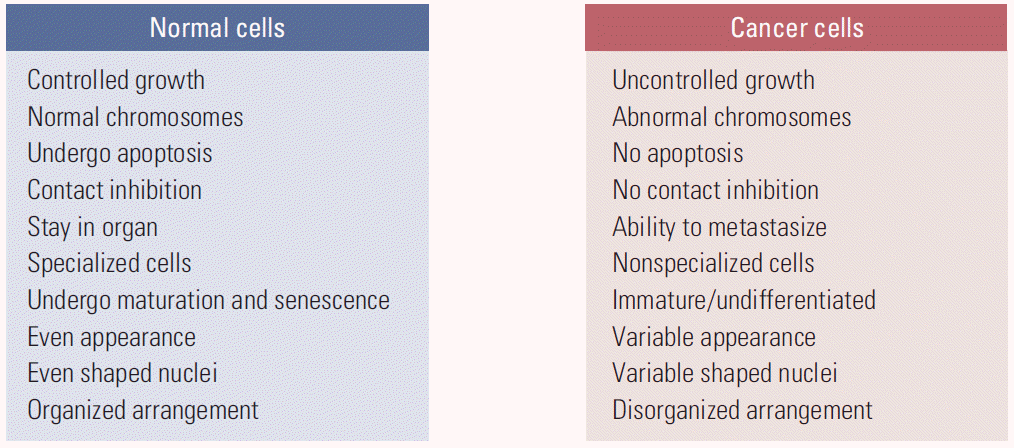
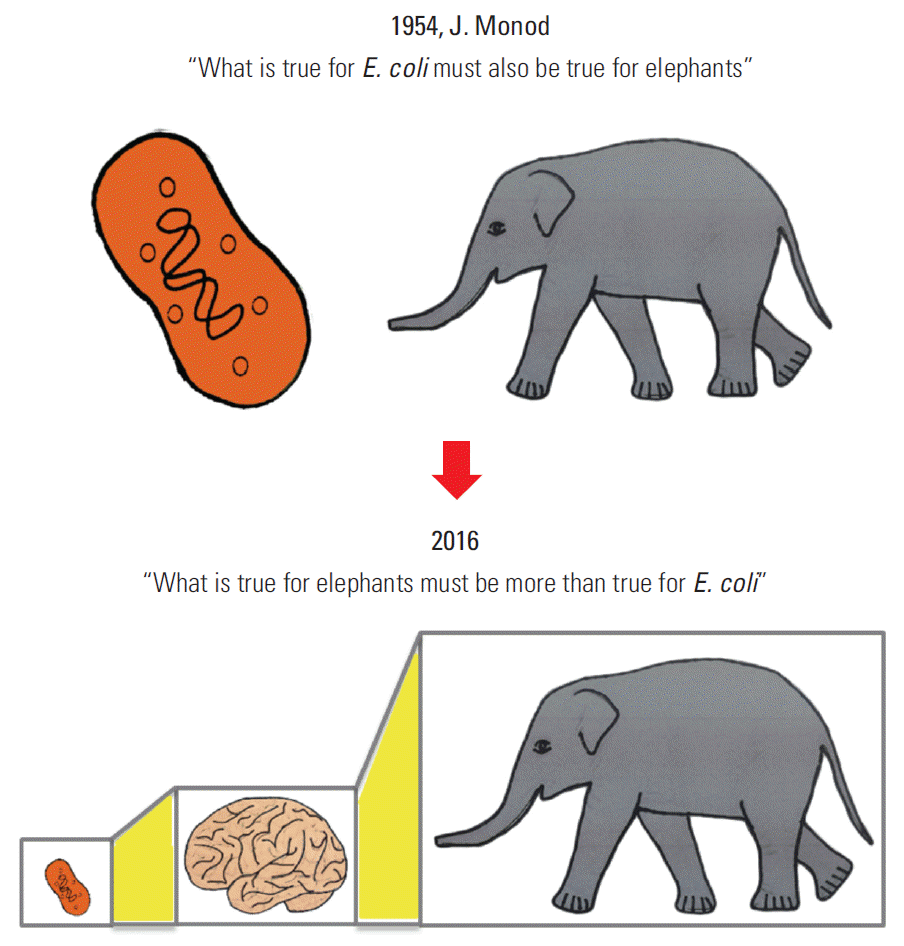
“What is true for E. coli must also be true for elephants.” This quote by J. Monod in 1954, when molecular biology studies began in earnest, is still valid today at the molecular level. Since then, research in biological sciences has centered on the cell and cellular molecules, contributing to the current focus of the life sciences on proteins and genes at the molecular level of a single cell. Accordingly, cancer research has concentrated on cancer cells, and the differences between normal cells and cancer cells, including their genetic variations (Fig. 1) [1].
For example, R. A. Weinberg’s “A perspective on cancer cell metastasis,” published in 2011, focused on cancer cells by explaining that the cancer overcomes the six steps of metastasis via the capability gained by the cancer cell [2]. Therefore, the current cancer research focuses on various functions of proteins and genes at the molecular or single-cell level without considering the cell-surrounding environment and the interaction between cells. As a result, development of anticancer drugs has been based on the assumption that all cancer cells share a certain set of characteristics during abnormal growth. This principle underpinned the development of drugs with anti-proliferative effects, starting with the alkylating agents in 1946 [3]. The search for a standard treatment for all cancers was launched through the development of such cytotoxic anticancer drugs.
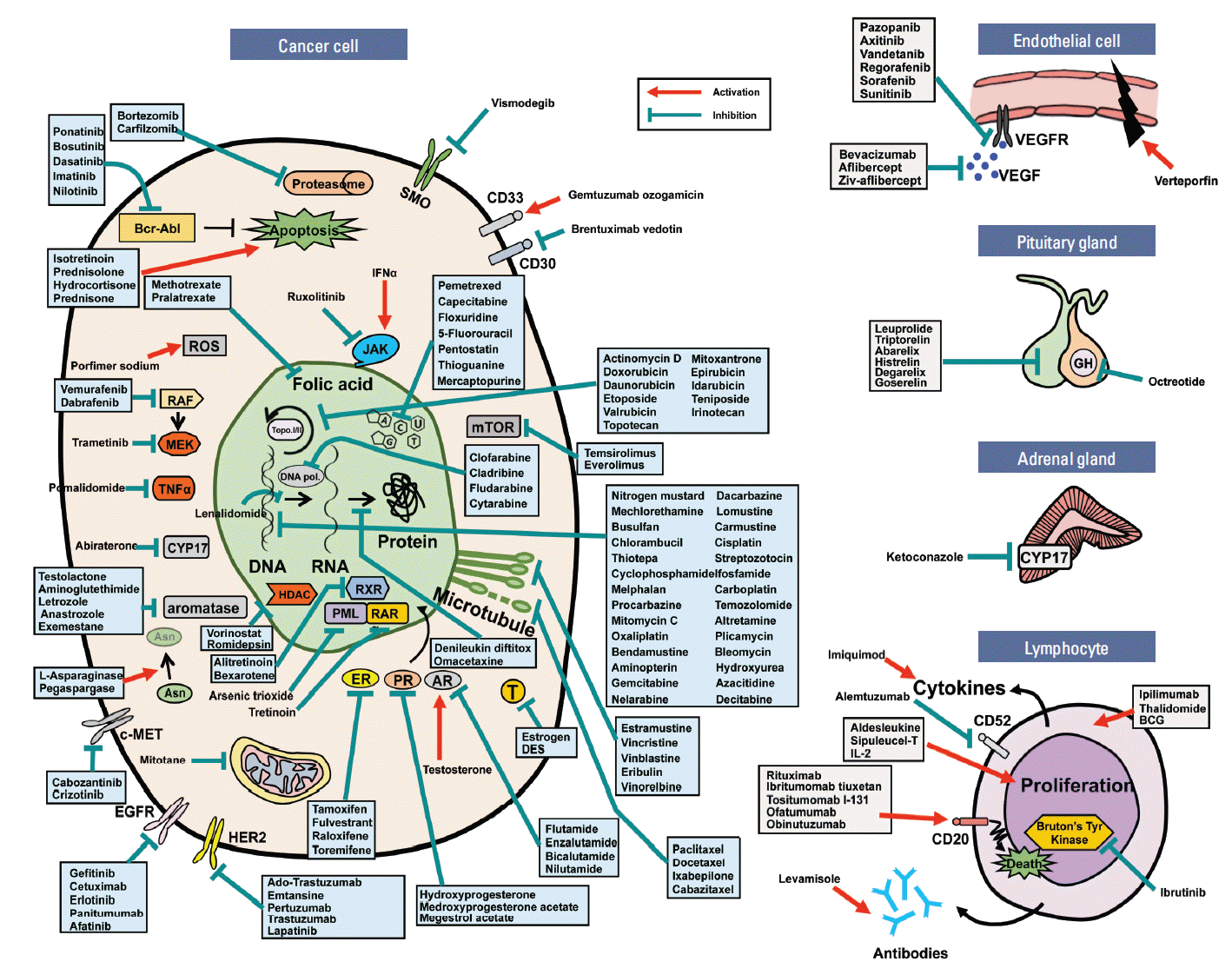
Accordingly, most contemporary anticancer drugs inhibit cell division. These cytotoxic anticancer drugs effectively suppress the division of both cancer and normal cells by blocking the general mechanism of cell division, leading to a multitude of side effects [1]. However, recently developed agents with molecular targets—for example, signaling factors— are able to distinguish normal cells by targeting signal transduction pathways uniquely related to cancer cell division and attempting to normalize its function [1,4-6]. Such agents target the specific abnormal signaling factors in cancer cells rather than the general targeting strategy associated with previous anticancer drugs. Among the target sites of the 160 anticancer drugs approved by Food and Drug Administration (United States), up to 80% are focused on cancer cells, and most of the targets are metabolic pathways and signal transduction pathways related to cancer cell division, as shown in Fig. 2. The remaining 20% of anticancer drugs target other components, such as immune cells and endothelial cells. A detailed look at 80% of anticancer drugs shows that alkylating agents act directly on DNA by suppressing cell proliferation, while anti-metabolites act on the biosynthesis of nucleic acids, such as DNA and RNA, thereby leading to apoptotic cell death [7,8]. In addition, hormonal agents act on intracellular hormone receptors, such as estrogen or progesterone receptors, whereas plant alkaloids and antibiotics mainly act on microtubules and DNA topoisomerase. Many of the recently developed targeted agents block the aberrantly activated cell surface receptor tyrosine kinase, such as epidermal growth factor receptor and human epidermal growth factor receptor 2 [9,10]. The targeted anticancer drugs for these proteins were developed based on technologies such as monoclonal antibodies, DNA sequencing, and polymerase chain reaction, which were intensively developed in the 1970s to the 1980s [1,11].
However, these anticancer drugs were not as successful as expected for most solid tumors, and this was discussed at the World Oncology Forum held in Lugano, Switzerland in 2012 [12]. Current anticancer drugs, including targeted drugs, are not very effective against advanced malignant tumors other than hematological tumors [12]. To overcome the limitations of these current cancer drugs, we must reevaluate current cancer research.
Go to : 
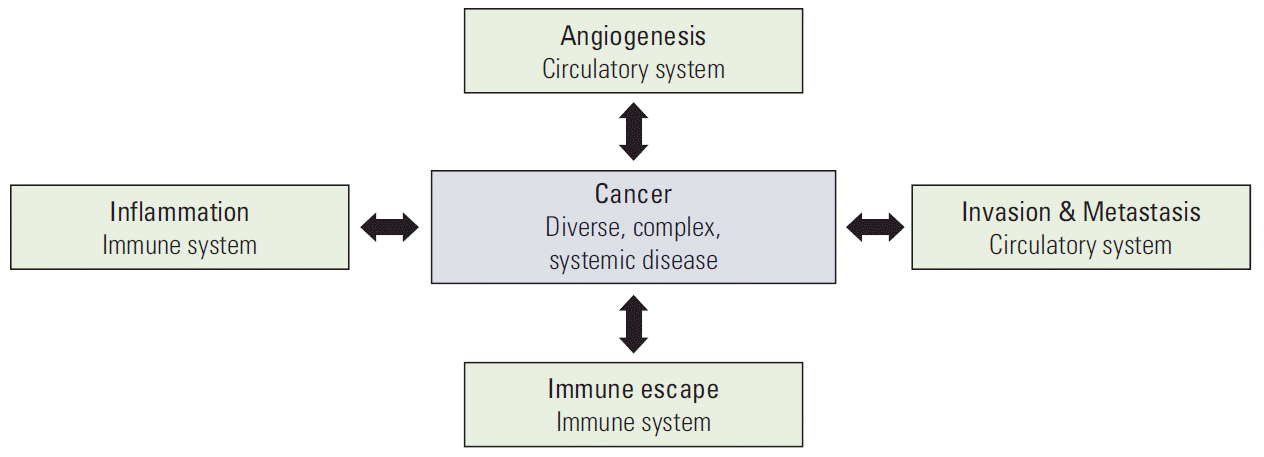
Expanded knowledge today indicates that multicellular organisms rely on biological phenomena that are non-existent in unicellular organisms. Multicellular life phenomena exist in tissues or organs that are aggregates of cells and composed of specialized systems, such as the circulatory system, nervous system, and immune system. Defects and functional disorders of these systems are closely related to the diseases of multicellular organism. Based on these principles, the understanding of physiological phenomena has changed over the last 60 years, as shown in Fig. 3. In this regard, cancer can also be considered as a systemic disorder, because malignant cancer causes human death via metastasis through the vascular and lymphatic systems in its advanced stage (Fig. 4).
Multicellular organisms, such as humans, exhibit the emergence of new life phenomena at a superior level through hierarchical systematization (cell-tissue-organ-organism), unlike unicellular organisms. Vertebrates, including humans, strictly rely on the cooperative interaction between various tissues and organs that perform specialized functions to maintain their vital activities (Figs. 3 and 5). Each tissue performs its unique function through the organic interaction of its various component cells, including specialized cells working in a specific tissue. For example, recent studies have shown that high-level brain functions, such as learning, memory and creativity, as well as various cerebral disorders, such as Alzheimer’s disease and Parkinson’s disease, are mediated by the complex cellular interaction network consisting of nerve cells, glial cells, astrocytes, and endothelial cells rather than by a specific protein or gene [13-16]. Similarly, tumor cells become malignant by interacting with various surrounding cells during tumorigenesis [17], and it has been shown that the tumor microenvironment, including the surrounding cells, plays an important role in tumorigenesis [18,19] (Fig. 4).
Accordingly, the new challenge to the life sciences is the establishment of a new perspective and methodology for studying the high-level life phenomena present in such tissues and organs. Thus, the cellular network at the meta-molecular level should become the focus of studies using an integrative approach, drawing upon what was learned from the previous reductive approaches and using changes in the paradigm to study tissue characteristics and functions from the superior level of cells. The cellular level shows a high specificity from the numerous activities accumulated at the molecular level, and thus an understanding of diseases drawn from previous research focused on only molecules is not sufficient to understand systemic diseases. Consequently, new alternatives to overcome these limitations could be provided by studying at the cell network level.
From this point on, studies explaining the interaction network between tissue-specific cells in various tissues and the emergence of tissue specificity because of this cell network are required, as shown in Fig. 5.
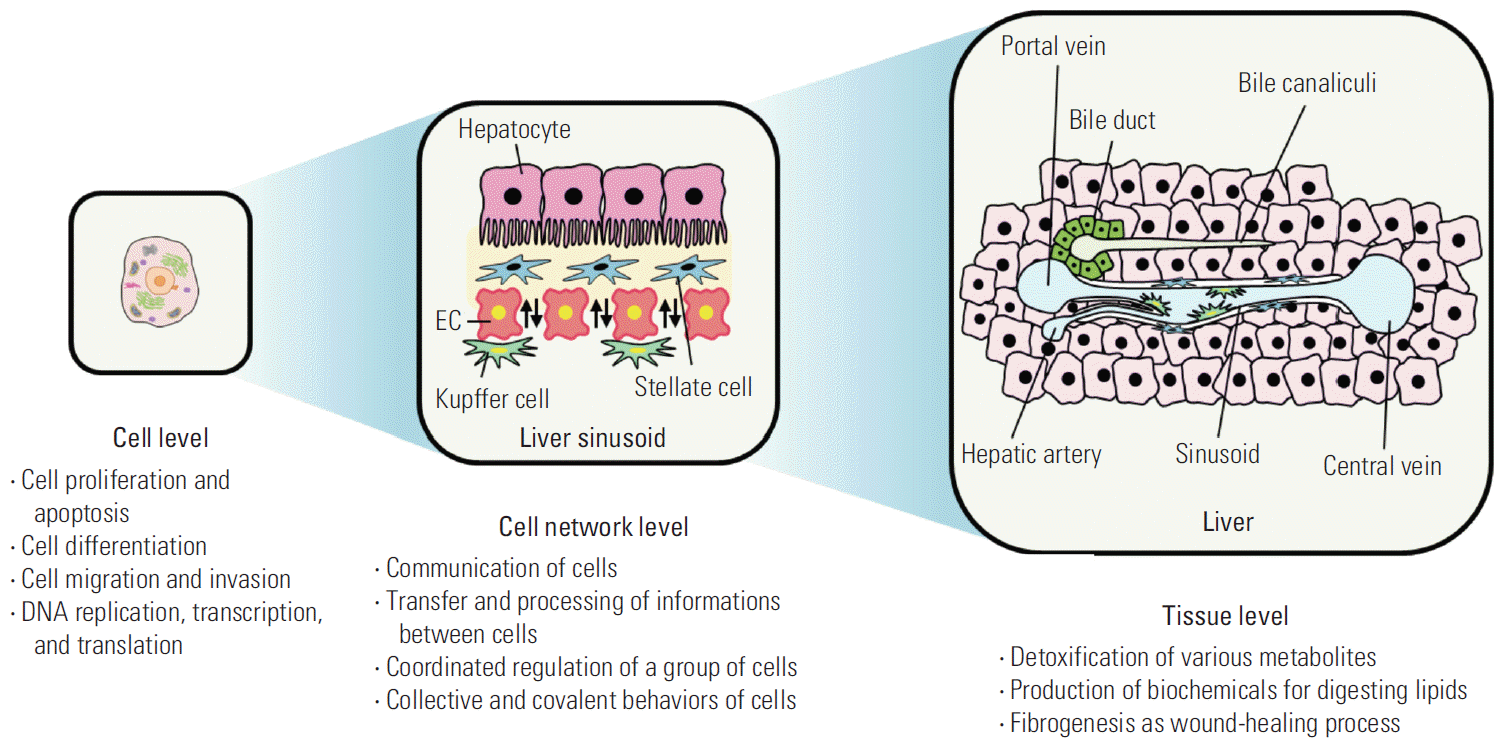
The tissue-specific cell network plays an important role in integrating and expressing tissue characteristics and functions via unique interactions among various cells [20]. This function differs from division, differentiation, and migration of individual cells at the cellular level; rather, it involves a synchronized coordination in the migration, division, and differentiation and the spatiality of various cellular functions of a group of cells. That is, a cell group constitutes a tissue-specific cell network and collectively and covalently shares the division and migration simultaneously at the entire tissue level. This aspect presents a new field of interest and study at the tissue-specific cell network level (Fig. 5). The types of tissue-specific cell networks in the body are poorly understood at present.
A typical example of a tissue-specific cell network is the network of blood vessels. Each human blood vessel consists of endothelial cells (ECs), but the same ECs create a completely different tissue-specific vascular system in terms of function and characteristics, depending on which tissue-specific cells form the network. Representative examples are the blood-brain barrier (BBB) in brain tissue and the sinusoid in liver tissues (Fig. 6). The BBB can transfer nutrients, such as glucose and amino acids, from blood, but it strictly blocks toxic materials to protect the nervous system of the brain. Thus, cerebrovascular ECs have the strongest tight junctions in the human body and various transport systems [21,22]. During the BBB developmental process, brain-specific astrocytes form a network with ECs and induce strong tight junctions between ECs [23]. However, liver tissues form fenestrae between ECs to loosen the cellular adhesion for the active exchange of materials between blood vessels and hepatocytes to supply nutrients to the body after detoxifying the materials at the space of Disse in the hepatic plate [24,25]. This creates a perforating vascular structure called a sinusoid, which allows the free exchange of materials. Perforating ECs can also be observed in the intestinal mucosa, endocrine gland, and glomerulus in the kidney and pancreas. A network with liver-specific surrounding cells is expected to be important in the development of perforating ECs, but detailed studies have not yet been performed.
Thus, new studies based on the cell network, unlike previous studies at the molecular level, can elucidate how cell networks specifically generate tissues and can determine the tissue-specific functions. These studies can be expanded to introduce a methodology for interpreting the pathogenesis of systemic disorders such as cancer in a novel manner. Currently, few cell networks are known to exist in various human tissues, and thus, various cell networks must be first investigated, and the emergence of new structures and functions at the tissue level via the cell network can then be studied in detail. Subsequently, damage and destruction of the cell network and its recovery during the pathogenesis of disorders such as cancer should be studied so that a new perspective can be proposed on the pathogenesis and treatment of systemic disorders.
More than 10 years ago, we noted that the research of neural cell and blood vessel should be re-scoped from individual molecular level to working group of cell network, which means the minimal working unit between neural cells and vessels and now named as the neurovascular unit [20,26,27]. For the last 10 years, the scope of cancer research also has been enlarged from the molecules of individual cancer cells to the interaction of cancer cells and other surrounding normal cells in the concept of tumor microenvironment [17-19]. The presence of other normal cell, such as cancer-associated fibroblast, in tumors were identified and also the interaction between these normal cells and cancer cells are being unveiled [28]. In addition, the cancer cells take advantages of the normal immune cells for their survival and evading the host defense system [29,30]. We now realized the limit of molecular study in the cells and changed our view to the interaction of cancer cells to other cells. Therefore, these recently accumulated ampules of results in the tumor microenvironment may compensate our limit of understanding of cancer, in part. However, still we are only looking at the very limited parts of cell network in the tumor tissue. In addition, virtually most of these studies aim to explain the complicate systems with small number of molecules in the small population of cells.
Go to : 
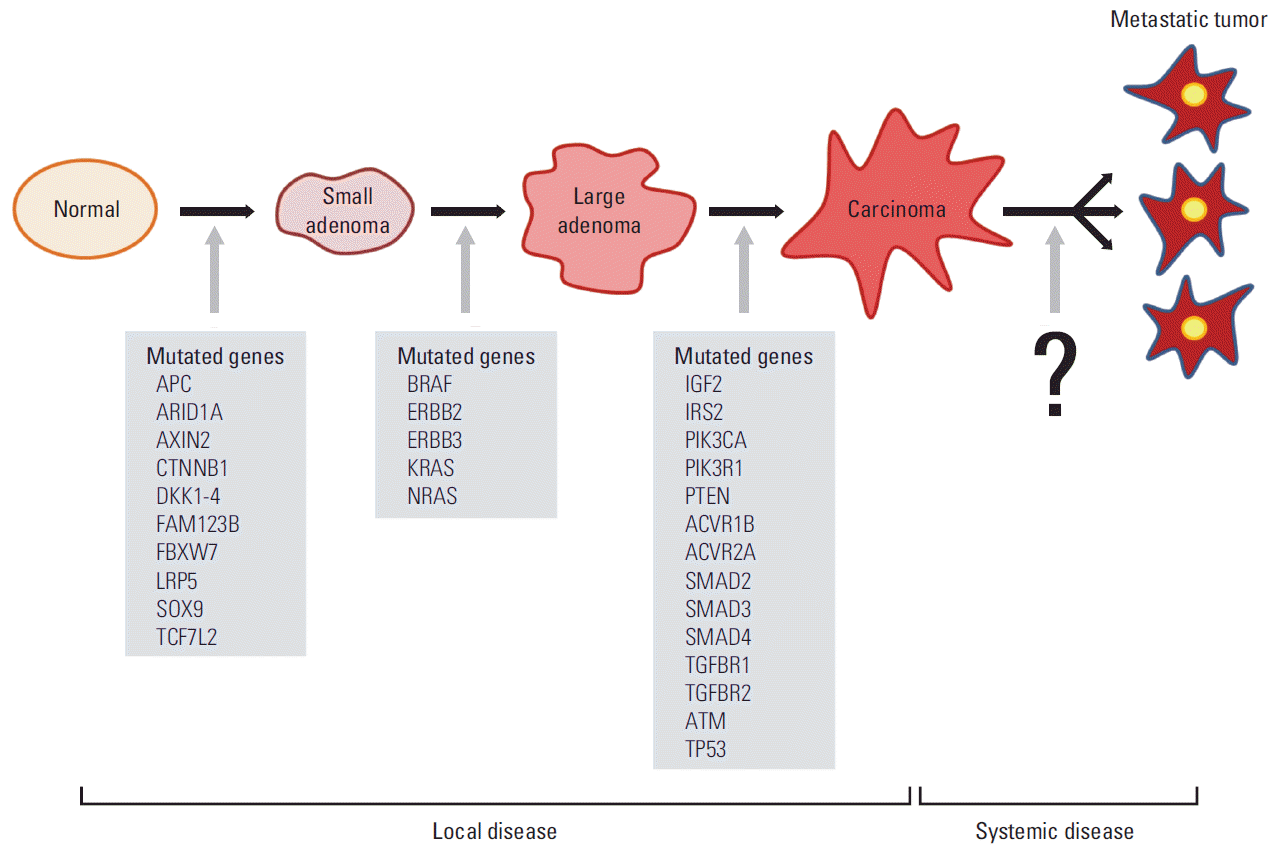
Recent cancer genomic studies allow the large-scale screening of genetic mutations for various human cancers not in animal tissues or in cancer cell lines [31,32]. The resulting genomic mutations are shown in Fig. 7 during the transformation of normal tissues to benign tumors and of benign tumors to malignant tumors [32-34]. However, the genetic mutations that are consistently common during the transformation of non-metastatic tumors to metastatic tumors have not yet been defined [32,35]. This result strongly suggests the importance of the interaction network between cancer cells and their surrounding cells in the advanced stage. Therefore, in the tumorigenesis, the step from normal epithelial tissue to adenoma, and then to carcinoma in situ, can potentially be regarded as a local disease predominantly characterized by cell proliferation, whereas the final step from carcinoma to the metastatic tumor is presumed to involve a change to a systemic disease. Thus, the intercellular interaction network of cancer tissues is expected to play a critical role during this change of cancer, from a local abnormality to a systemic disease. Therefore, investigation of the tumor-specific cell network can propose a new breakthrough in explaining the metastatic stage.
Go to : 
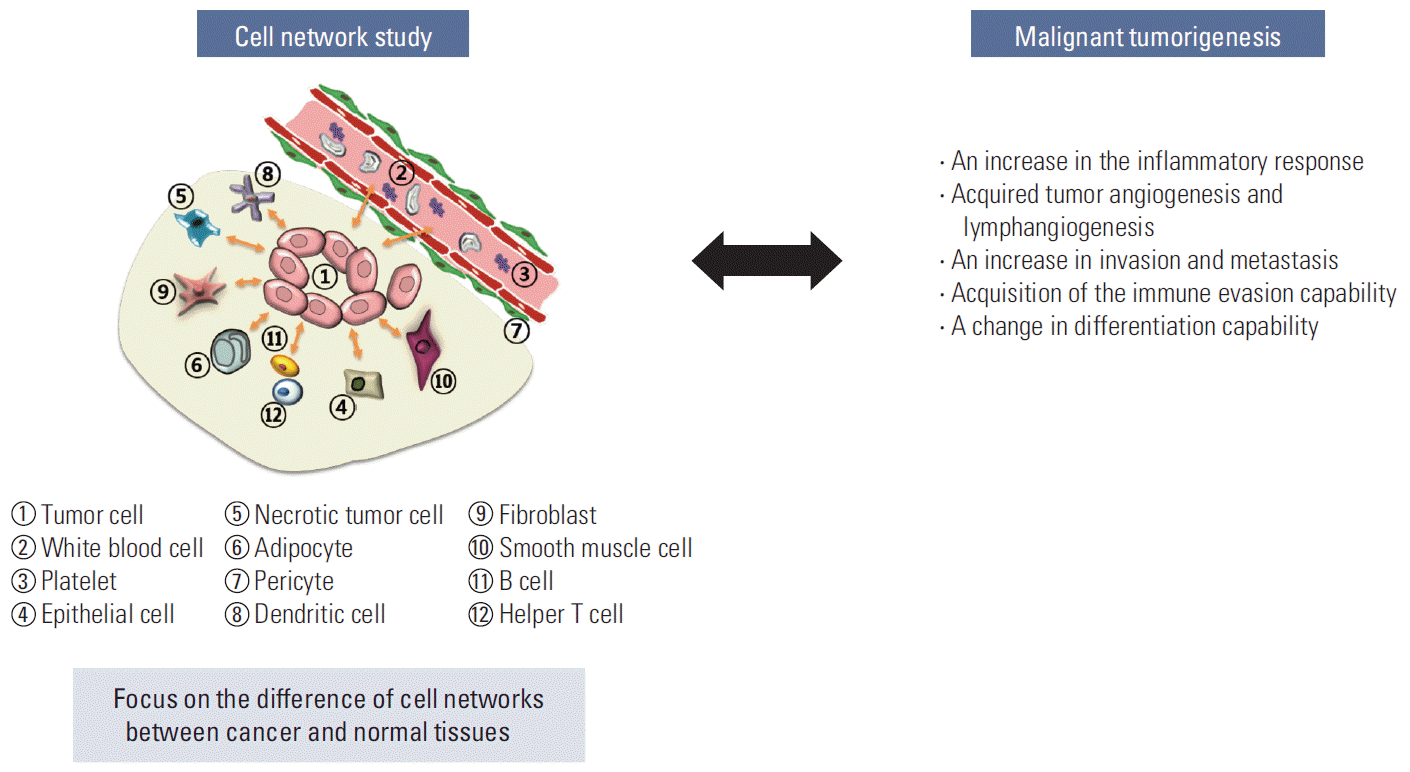
If the molecular changes in the interacting cells are the point of focus in the cell network research, it would be more difficult to identify a solution. The cell network study of cancer tissue becomes enigmatic beyond imagination considering that we now understand the dynamic heterogeneity of cancer cells. The current massive molecular level study on these heterogeneous populations will generate a huge amount of data and information, which will be too complex to solve the problems. The intratumoral, ever-changing heterogeneity is another big huddle making it more complicate but we ignore now. Such complexity can be overcome by directly relating the tumor-specific cell network and the malignant tumorigenesis instead of investigating them at the molecular level of the cells. The research targets of the tumorspecific cell network may include an increase in the inflammatory response, an increase in invasion/metastasis, acquisition of the immune evasion capability, a change in the differentiation capability, etc as shown in Fig. 8. Through these attempts, tumorigenesis can be newly understood and defined at the cell network level, which would enable the development of new-concept cancer drugs and treatments that can regulate or block malignant metastatic tumorigenesis. It is time to study the tumor-specific cell networks, not cancer cells.
Go to : 
ACKNOWLEDGMENTS
This study was supported by the Global Core Research Center (GCRC) Program (2011-0030001) funded by the Korean Ministry of Science, ICT and Future Planning (MSIP) through the National Research Foundation (NRF).
Go to : 
References
1. Kim KW, Rho JK, Wee HJ, Kim C. Cancer drug discovery: science and history. Dordrecht: Springer;2016.
5. Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009; 361:947–57.

6. Drugs of choice for cancer. Treat Guidel Med Lett. 2003; 1:41–52.
7. Mohamed MF, Mohamed MS, Fathi MM, Shouman SA, Abdelhamid IA. Chalcones incorporated pyrazole ring inhibit proliferation, cell cycle progression, angiogenesis and induce apoptosis of MCF7 cell line. Anticancer Agents Med Chem. 2014; 14:1282–92.

8. Lord RM, Hebden AJ, Pask CM, Henderson IR, Allison SJ, Shepherd SL, et al. Hypoxia-sensitive metal beta-ketoiminato complexes showing induced single-strand DNA breaks and cancer cell death by apoptosis. J Med Chem. 2015; 58:4940–53.
9. Goel S, Wang Q, Watt AC, Tolaney SM, Dillon DA, Li W, et al. Overcoming therapeutic resistance in HER2-positive breast cancers with CDK4/6 inhibitors. Cancer Cell. 2016; 29:255–69.

10. Brack S, Attinger-Toller I, Schade B, Mourlane F, Klupsch K, Woods R, et al. A bispecific HER2-targeting FynomAb with superior antitumor activity and novel mode of action. Mol Cancer Ther. 2014; 13:2030–9.

11. Liu JK. The history of monoclonal antibody development: progress, remaining challenges and future innovations. Ann Med Surg (Lond). 2014; 3:113–6.
13. Compston A, Zajicek J, Sussman J, Webb A, Hall G, Muir D, et al. Glial lineages and myelination in the central nervous system. J Anat. 1997; 190(Pt 2):161–200.
14. Corty MM, Freeman MR. Cell biology in neuroscience: architects in neural circuit design: glia control neuron numbers and connectivity. J Cell Biol. 2013; 203:395–405.
16. Mosley RL, Hutter-Saunders JA, Stone DK, Gendelman HE. Inflammation and adaptive immunity in Parkinson's disease. Cold Spring Harb Perspect Med. 2012; 2:a009381.

17. Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009; 9:239–52.

18. Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013; 19:1423–37.

19. Mao Y, Keller ET, Garfield DH, Shen K, Wang J. Stromal cells in tumor microenvironment and breast cancer. Cancer Metastasis Rev. 2013; 32:303–15.

20. Park JA, Choi KS, Kim SY, Kim KW. Coordinated interaction of the vascular and nervous systems: from molecule- to cellbased approaches. Biochem Biophys Res Commun. 2003; 311:247–53.

21. Schlosshauer B. The blood-brain barrier: morphology, molecules, and neurothelin. Bioessays. 1993; 15:341–6.

22. Lee SW, Kim WJ, Choi YK, Song HS, Son MJ, Gelman IH, et al. SSeCKS regulates angiogenesis and tight junction formation in blood-brain barrier. Nat Med. 2003; 9:900–6.

23. Obermeier B, Daneman R, Ransohoff RM. Development, maintenance and disruption of the blood-brain barrier. Nat Med. 2013; 19:1584–96.

24. Collardeau-Frachon S, Scoazec JY. Vascular development and differentiation during human liver organogenesis. Anat Rec (Hoboken). 2008; 291:614–27.

25. Mescher AL. Junqueira’s basic histology. New York: McGraw-Hill;2010. p. 212–33.
27. Lo EH, Broderick JP, Moskowitz MA. tPA and proteolysis in the neurovascular unit. Stroke. 2004; 35:354–6.

28. Silzle T, Randolph GJ, Kreutz M, Kunz-Schughart LA. The fibroblast: sentinel cell and local immune modulator in tumor tissue. Int J Cancer. 2004; 108:173–80.

29. Fukuda K, Kobayashi A, Watabe K. The role of tumor-associated macrophage in tumor progression. Front Biosci (Schol Ed). 2012; 4:787–98.
31. Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008; 455:1061–8.
32. Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer genome landscapes. Science. 2013; 339:1546–58.

33. Cancer Genome Atlas Research Network, Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013; 45:1113–20.

34. Tomczak K, Czerwinska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn). 2015; 19:A68–77.
Go to : 




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download