Abstract
Purpose
Papillary thyroid carcinomas (PTCs) frequently involve genetic alterations. The objective of this study was to investigate genetic alterations and further explore the relationships between these genetic alterations and clinicopathological characteristics in a high-recurrence risk (node positive, N1) PTC group.
Materials and Methods
Tumor tissue blocks were obtained from 240 surgically resected patients with histologically confirmed stage III/IV (pT3/4 or N1) PTCs. We screened gene fusions using NanoString’s nCounter technology and mutational analysis was performed by direct DNA sequencing. Data describing the clinicopathological characteristics and clinical courses were retrospectively collected.
Results
Of the 240 PTC patients, 207 (86.3%) had at least one genetic alteration, including BRAF mutation in 190 patients (79.2%), PIK3CA mutation in 25 patients (10.4%), NTRK1/3 fusion in six patients (2.5%), and RET fusion in 24 patients (10.0%). Concomitant presence of more than two genetic alterations was seen in 36 patients (15%). PTCs harboring BRAF mutation were associated with RET wild-type expression (p=0.001). RET fusion genes have been found to occur with significantly higher frequency in N1b stage patients (p=0.003) or groups of patients aged 45 years or older (p=0.031); however, no significant correlation was found between other genetic alterations. There was no trend toward favorable recurrence-free survival or overall survival among patients lacking genetic alterations.
Papillary thyroid carcinoma (PTC) is the most common thyroid cancer, and its annual incidence is increasing worldwide [1]. Although PTCs are usually curable, with a 10-year survival rate of 80%-90%, the rate of disease recurrence or persistence is high. Recurrence occurs locoregionally in 5%-20% of patients and as distant metastasis in 10%-20% of patients during long-term follow-up after initial therapy [2]. Patients older than 45 years at the time of initial diagnosis have a much worse prognosis when recurrence occurs [3].
Recent understanding of the molecular pathogenesis of PTC has resulted from identification of genetic alterations in various signaling pathways. Genetic alterations in the mitogen-associated protein kinase (MAPK) pathway, such as BRAF point mutations, RAS point mutations, and RET rearrangements, play important roles in the initiation and progression of PTC [4]. BRAF mutations are the most common genetic alterations found in PTC, followed by RET rearrangements and RAS mutations. These mutations are found in more than 70% of PTCs and are almost always mutually exclusive [5,6]. However, recent studies have revealed concomitant mutations in advanced stages of PTC [3,7], although the relationship between genetic alterations and their influence on prognosis remains unclear.
BRAF mutations are the most common type of genetic alterations in PTC, with an incidence ranging from 28% to 83% and an overall rate of 45% [4,8]. More than 90% of all BRAF mutations consist of a valine-to-glycine substitution at codon 600 (V600E) in exon 15. Although the impact of BRAF mutations in PTC is incompletely defined, many studies have demonstrated a strong association of BRAFV600E with poor clinicopathological outcome of PTC [9,10]. The RAS gene family, including NRAS, HRAS, and KRAS, encodes the 21-kDa G-proteins, which influence the MAPK and phosphoinositide 3-kinase (PI3K) signaling pathways in thyroid cancer. Point mutations in codons 12, 13, and 61 of RAS have been found in thyroid cancer [4,11]; however, their reported frequency varies among studies [12]. Mutations at codon 61 of RAS cause reduction of GTPase activity and are related to aggressiveness of PTC [11]. Genetic alterations of the PIK3CA gene, specifically activating mutations in exons 9 and 20, have been found widely in human cancer. However, several studies have reported that PIK3CA mutation is uncommon in PTC [13].
A recent study by The Cancer Genome Atlas (TCGA) revealed mutually exclusive recurrent kinase fusions in thyroid cancer [14]. Rearrangements of RET are commonly seen in PTC [15], and the most common RET fusions are paracentric fusions, with the coiled-coil domain containing 6 (CCDC6) contributing ~80% and the nuclear receptor coactivator 4 (NCOA4) contributing ~10% of all known RET rearrangements [4]. RET fusions result in ligand-independent dimerization and constitutive RET activation. Thus, RET fusions are classical oncogenes that activate the MAPK and PI3K signaling pathways. Recurrent fusions involving members of the neurotrophic tyrosine receptor kinase (NTRK) family have also been identified in PTC [16]. Although NTRK gene rearrangements are less common than RET rearrangements, they play direct roles and represent an early event in the process of thyroid carcinogenesis [16]. The paired box 8 (PAX8)–peroxisome proliferator activated receptor-γ (PPARγ) fusion gene is another prominent recombinant oncogene that has been implicated in thyroid cancer. The PAX8-PPARγ fusion gene is most commonly found in follicular thyroid cancer, i.e., follicular variant PTC [4,12]. However, this gene has a low frequency in Asian populations [12].
There is a greater incidence of PTC in Korea than in other countries, and this increased prevalence is accompanied by differences in the clinicopathological characteristics of these tumors [17]. Although numerous studies have attempted to identify prognostic markers that distinguish high-recurrence risk PTCs, the usefulness of genetic analysis in PTC patient management is still uncertain [8,12,18]. In the present study, we investigated genetic alterations in high-recurrence risk PTC from Korea and their association with various clinicopathological characteristics by conducting a NanoString nCounter gene fusion assay and direct sequencing of seven hotspot mutations in BRAF, KRAS, or PIK3CA. These fusion genes and hotspots were selected because they are associated with aggressive features of various epithelial cancers, including thyroid cancer.
From January 2004 to August 2008, a total of 855 patients underwent surgery (subtotal or total thyroidectomy or neck dissection) for primary thyroid carcinoma at Samsung Medical Center (SMC, Seoul, Korea). Of these patients, we selected those with histologically confirmed T3 to T4 or N1 PTC. Histopathological diagnoses according to the World Health Organization (WHO) classification system were obtained from pathological reports. We excluded histological variants or mixed-type PTC. After application of our selection criteria, a total of 240 patients were included in this study as the high-recurrence risk PTC group. The diameter of tumors ranged from 0.4 to 10.5 cm, with a median size of 1.5 cm. The tumors of 40.8% of the patients (98 cases) showed multifocality and 94.6% of patients (227 cases) presented extrathyroid invasion. Moreover, all patients were found to have lymph node metastases (range, 1 to 41; median, 4) at the time of initial surgery. A pathologist reviewed the tumor specimens and histologic reviews of all samples, and thyroid tumor tissue blocks were obtained from these patients. Total RNA and DNA of sufficient quality were successfully extracted from all 240 specimens available. Clinical data describing the surgical procedures performed, intraoperative findings, histopathologic data, and preoperative and postoperative status of patients were retrospectively collected and reviewed (from January 2004 to December 2014). The median follow-up duration was 95.8 months (range, 0.9 to 130.4 months). This study was approved by the Institutional Review Board of Samsung Medical Center.
Direct sequencing of the BRAF (exon 15), KRAS (exons 2 and 3), and PIK3CA (exons 9 and 20) genes was conducted. Briefly, genomic DNA was extracted from formalin-fixed, paraffin-embedded (FFPE) tissue sections using a QIAamp DNA FFPE Tissue Kit (Qiagen, Valencia, CA). Purified DNA was quantified using NanoDrop (Invitrogen Life Technologies, Carlsbad, CA) and Qubit (Invitrogen Life Technologies). The BRAF (V600), KRAS (G12, G13, and Q61), and PIK3CA (E542, E545, and H1047) hotspot mutations were evaluated by polymerase chain reaction, followed by Sanger sequencing using a BigDye Terminator v3.1 Cycle Sequencing (Applied Biosystems, Foster City, CA) on an ABI 3730XL automated sequencer (Applied Biosystems).
Total RNA was isolated from two to three FFPE tissue sections (10 μm thick) using an miRNeasy FFPE Kit (Qiagen) according to the manufacturer’s instructions. The probe sets were custom designed and synthesized by NanoString Technologies (Seattle, WA), and nCounter assays were performed according to the manufacturer’s protocol. Briefly, 500 ng of total RNA was hybridized to nCounter probe sets for 16 hours at 65°C. Samples were then processed using an automated nCounter Sample Prep Station (NanoString Technologies). Cartridges containing immobilized and aligned reporter complexes were subsequently imaged on an nCounter Digital Analyzer (NanoString Technologies). Reporter counts were collected using the NanoString’s nSolver analysis software ver. 1, normalized, and analyzed.
Overall survival (OS) was calculated from the date of diagnosis to the date of death or final follow-up. Relapse-free survival (RFS) was defined from the date of first surgery until tumor progression, death, or end of follow-up. Kaplan-Meier methodology was used to estimate survival probabilities, which were expressed as the mean with the range and two-sided 95% confidence interval (CI) and compared between two or more groups of patients using the log-rank test. Differences between the clinicopathologic characteristics of patients with high-risk PTC and various genetic alterations were evaluated using either Pearson’s chi-squared test or Fisher exact test. A two-sided p-value < 0.05 was considered statistically significant. All analyses were performed using SPSS ver. 18.0 (SPSS Inc., Chicago, IL).
Among 855 patients with PTC whose data were obtained from January 2004 to August 2008, 240 (28.1%) had pathological stage T3/4 or N1. These patients were designated as the high-recurrence risk PTC group. The general characteristics of the study population are summarized in Table 1. The median patient age was 46 years, and 73% were female. Total or subtotal thyroidectomy was performed on 236 patients and 175 patients, respectively, with neck dissection of the involved compartments performed for clinically apparent or biopsy-proven lymph node metastasis. Four patients underwent dissection of deep cervical lymph nodes. All but three patients (i.e., 237 of 240) received postoperative radioactive iodine ablation therapy.
To detect the BRAFV600E mutation, exon 15 of BRAF was sequenced. The BRAFV600E mutation was observed in 190 of 240 patients (79.2%). No KRAS mutation was detected at G12 or G13 in exon 2 or Q61 in exon 3. PIK3CA mutations were found in 172 patients (71.7%), with one (0.4%) E542K and 24 (10%) E545A mutations in exon 9; however, no mutation was noted at H1047 in exon 20. We identified 138 patients (57.7%) with concomitant BRAFV600E and PIK3CAE545A mutations (Fig. 1).
RET fusions, which were the most frequent mutations, were detected in 24 patients (10.0%). Among these, 17 patients (7.1%) had an RET exon 12 fusion with CCDC6, NCOA4, or PRKAR1A. ETV6-NTRK3 fusions were found in five patients (2.1%), three with ETV6-NTRK3 (E4-N14) fusion (1.3%), and two with ETV6-NTRK3 (E5-N15) fusion (0.8%). TMP3-NTRK1 (T7-N10) fusion was found in one patient (0.4%). No EML4-ALK or PAX8-PPARγ fusions were detected. All fusions were mutually exclusive.
Among the 240 patients, 207 (86.3%) showed at least one genetic alteration. A total of 36 patients (15%) had two or more concomitant mutational events, with 34 (14.2%) having two events, and two patients (0.8%) having three events (Table 2). Both patients with PTC that had three events had concomitant BRAF mutation, PIK3CA mutation, and RET fusion.
PTCs harboring BRAFV600Emutations were associated with RET wild-type expression (p=0.001). RET fusion genes have been found to occur with significantly higher frequency in patients aged ≥ 45 years (p=0.031). However, no significant correlation was found with other genetic alterations (Table 3).
The overall median follow-up time was 95.8 months (range, 0.9 to 130.4 months) after initial treatment. During the follow-up period, 32 patients (13.3%) experienced relapse and 15 (6.3%) died (Table 1). There were significant differences in RFS between the two stage categories of the American Joint Committee on Cancer/Tumor-Nodes-Metastasis (AJCC/TNM) stage categorization system (hazard ratio [HR], 0.3; 95% CI, 0.15 to 0.63) (Fig. 2A). In addition, there was a trend toward short RFS in the high-risk patient group relative to the intermediate-risk groups as defined by American Thyroid Association (ATA) risk stratification (HR, 0.3; 95% CI, 0.08 to 1.19) (Fig. 2B). To determine if concomitant genetic alterations were associated with poor clinical outcome, we analyzed the RFS or OS of patients with concomitant alterations versus those who had only one or no alteration. Kaplan-Meier analysis revealed no significant differences in RFS or OS among patients with no alterations, a single alteration, two alterations, and three alterations, although patients with no genetic alterations had shorter RFS than those with more than one genetic alteration (Fig. 3).
Focusing on favorable prognosis of PTC, we examined a specific group of thyroid carcinomas with high recurrence risk and analyzed mutation status and gene fusions from a cohort of 240 patients. We found the simultaneous presence of BRAF and PIK3CA mutation in 20 patients (8.3%). Although concomitant genetic alterations occurred frequently in patients with high-recurrence risk PTC, these had no significant effect on clinical outcome.
The occurrence of BRAFV600E mutation in PTC has been extensively investigated. The frequency of BRAFV600E mutation in PTC varies from 18% to 90% in reported studies. A majority of the patients in our cohort (79.2%) had the BRAFV600E mutation, which is consistent with populations presented in the literature [9,19-21]. However, the prognostic value of the BRAFV600E mutation in PTC remains controversial. Several studies have found an association between BRAFV600Emutation and poor prognosis [10,22]. However, in other studies, the presence of BRAFV600E did not always seem to be associated with disease recurrence or mortality [23]. In the present study, the association between BRAFV600E and poor prognosis could not be validated because of the extremely high rate of the BRAFV600E mutation and the high prevalence of concomitant genetic alterations in our cohort.
The frequency of RAS mutation in thyroid cancer varies according to tumor cell origin, ranging from 0% to 57% [24]. Although RAS mutations are the second most commonly identified genetic alteration in thyroid cancer, they are primarily found in follicular-patterned tumors. In this study, KRAS mutation was not detected, suggesting that follicular variants of PTC may not have been included in our cohort.
The PIK3CA mutation has been examined in various differentiated thyroid cancers but has been found to be uncommon in this type of cancer [13]. Mutation in PIK3CA has mainly been found in poorly differentiated thyroid cancer or anaplastic thyroid cancer [25]. Of note, we observed a high incidence (10%) of PIK3CA mutation in high-recurrence risk PTC. Although COSMIC data showed that the most common PIK3CA hotspot mutation in PTC is PIK3CAE542K in exon 9, we found only one patient with that mutation. Instead, 24 of 240 patients (10%) had PIK3CAE545A in exon 9. This mutation has been reported in follicular thyroid carcinoma and the cribriform-morular variant of PTC [26]. The PIK3CAE545A mutation has been reported to induce AKT phosphorylation and possesses strong oncogenic potential in thyroid cancer [26]. Further research is necessary to determine the clinical significance of the PIK3CAE545A mutation in high-recurrence risk PTC. To date, several PI3K inhibitors have been developed and are being actively employed in clinical trials for a variety of solid tumors. Unfortunately, no specific predictive biomarker associated with these agents has been validated.
This study is unique in that it identified diverse translocation partners of known oncogenes, namely RET, NTRK1/3, ALK, and PPARγ, using a gene fusion detection assay. RET fusions were detected in 24 patients (10%), and NTRK1/3 fusions were found in six patients (2.5%). Rearrangements of RET are common in PTC and have been shown to play a role in disease pathogenesis. There are 13 different types of RET fusion, as determined by the types of partner genes, the most common being RET/PTC1 (CCDC6-RET) and RET/PTC3 (NCOA4-RET) [4]. These rearrangements result in constitutive tyrosine kinase activity of RET and activate the MAPK and PI3K-AKT pathways. To date, two multi-kinase inhibitors with RET tyrosine kinase inhibitor activity, vandetanib and cabozantinib, have been approved for the treatment of locally advanced and metastatic medullary thyroid carcinoma, and trials of additional multi-kinase inhibitor in thyroid cancers are in progress [27].
Consistent with previous studies [14,16], we observed a 2.5% incidence of NTRK1 and NTRK3 fusion in high-recurrent risk PTC. Several NTRK inhibitors, including AZD7451, LOX-101, and RXDX-101, have been developed, and early phase clinical trials are ongoing (ClinicalTrials.gov identifiers NCT02576431, NCT02097810, and NCT02568267). These gene rearrangements were mutually exclusive, but concomitant with other mutations. In advanced thyroid cancer, multiple genetic alterations may occur in different parts of the tumor, resulting in concomitant alterations. Concomitant subclonal genetic alterations, which are involved in tumor progression, were found in PTC [28,29]. Further studies are needed to determine the significance of these concomitant genetic alterations.
Thirty-three samples in our cohort had no detectable alteration in any genes analyzed. Given that potential limitations may exist with respect to the sensitivity of the detection method used, and that only potentially actionable genetic abnormalities were analyzed, it is uncertain whether these represent false- or true-negatives. Although no statistically significant differences were observed in RFS and OS in patients without any genetic abnormalities compared to those with genetic aberrations, there was a trend toward shorter RFS in this group, suggesting the existence of other undetected genetic alterations.
In conclusion, we found two or three concomitant genetic alterations in 36 patients. These results indicate intratumoral heterogeneity or multiclonal origins in the tumors. However, these concomitant genetic alterations did not affect RFS or OS, suggesting that the presence of multiple genetic alterations is not an indicator of tumor malignancy or of poor prognosis in high-recurrence risk PTC. Additional studies are necessary to identify genetic drivers in high-recurrence risk PTC patients lacking oncogenic alterations.
References
1. Ito Y, Nikiforov YE, Schlumberger M, Vigneri R. Increasing incidence of thyroid cancer: controversies explored. Nat Rev Endocrinol. 2013; 9:178–84.

3. Henderson YC, Shellenberger TD, Williams MD, El-Naggar AK, Fredrick MJ, Cieply KM, et al. High rate of BRAF and RET/PTC dual mutations associated with recurrent papillary thyroid carcinoma. Clin Cancer Res. 2009; 15:485–91.

4. Xing M. Molecular pathogenesis and mechanisms of thyroid cancer. Nat Rev Cancer. 2013; 13:184–99.

5. Soares P, Trovisco V, Rocha AS, Lima J, Castro P, Preto A, et al. BRAF mutations and RET/PTC rearrangements are alternative events in the etiopathogenesis of PTC. Oncogene. 2003; 22:4578–80.

6. Adeniran AJ, Zhu Z, Gandhi M, Steward DL, Fidler JP, Giordano TJ, et al. Correlation between genetic alterations and microscopic features, clinical manifestations, and prognostic characteristics of thyroid papillary carcinomas. Am J Surg Pathol. 2006; 30:216–22.

7. Costa AM, Herrero A, Fresno MF, Heymann J, Alvarez JA, Cameselle-Teijeiro J, et al. BRAF mutation associated with other genetic events identifies a subset of aggressive papillary thyroid carcinoma. Clin Endocrinol (Oxf). 2008; 68:618–34.

8. Hong AR, Lim JA, Kim TH, Choi HS, Yoo WS, Min HS, et al. The frequency and clinical implications of the BRAF(V600E) mutation in papillary thyroid cancer patients in Korea over the past two decades. Endocrinol Metab (Seoul). 2014; 29:505–13.
9. Kim TH, Park YJ, Lim JA, Ahn HY, Lee EK, Lee YJ, et al. The association of the BRAF(V600E) mutation with prognostic factors and poor clinical outcome in papillary thyroid cancer: a meta-analysis. Cancer. 2012; 118:1764–73.
10. Xing M, Alzahrani AS, Carson KA, Viola D, Elisei R, Bendlova B, et al. Association between BRAF V600E mutation and mortality in patients with papillary thyroid cancer. JAMA. 2013; 309:1493–501.

11. Benvenga S, Koch CA. Molecular pathways associated with aggressiveness of papillary thyroid cancer. Curr Genomics. 2014; 15:162–70.

12. Song YS, Lim JA, Park YJ. Mutation profile of well-differentiated thyroid cancer in Asians. Endocrinol Metab (Seoul). 2015; 30:252–62.

13. Xing M. Genetic alterations in the phosphatidylinositol-3 kinase/Akt pathway in thyroid cancer. Thyroid. 2010; 20:697–706.

14. Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nat Commun. 2014; 5:4846.

15. Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014; 159:676–90.
16. Greco A, Miranda C, Pierotti MA. Rearrangements of NTRK1 gene in papillary thyroid carcinoma. Mol Cell Endocrinol. 2010; 321:44–9.

17. Cho BY, Choi HS, Park YJ, Lim JA, Ahn HY, Lee EK, et al. Changes in the clinicopathological characteristics and outcomes of thyroid cancer in Korea over the past four decades. Thyroid. 2013; 23:797–804.

18. Chitikova Z, Pusztaszeri M, Makhlouf AM, Berczy M, Delucinge-Vivier C, Triponez F, et al. Identification of new biomarkers for human papillary thyroid carcinoma employing NanoString analysis. Oncotarget. 2015; 6:10978–93.

19. He G, Zhao B, Zhang X, Gong R. Prognostic value of the BRAF V600E mutation in papillary thyroid carcinoma. Oncol Lett. 2014; 7:439–43.

20. Cheng SP, Hsu YC, Liu CL, Liu TP, Chien MN, Wang TY, et al. Significance of allelic percentage of BRAF c.1799T > A (V600E) mutation in papillary thyroid carcinoma. Ann Surg Oncol. 2014; 21 Suppl 4:S619–26.
21. Li C, Lee KC, Schneider EB, Zeiger MA. BRAF V600E mutation and its association with clinicopathological features of papillary thyroid cancer: a meta-analysis. J Clin Endocrinol Metab. 2012; 97:4559–70.
22. Xing M, Alzahrani AS, Carson KA, Shong YK, Kim TY, Viola D, et al. Association between BRAF V600E mutation and recurrence of papillary thyroid cancer. J Clin Oncol. 2015; 33:42–50.

23. Henke LE, Pfeifer JD, Ma C, Perkins SM, DeWees T, El-Mofty S, et al. BRAF mutation is not predictive of long-term outcome in papillary thyroid carcinoma. Cancer Med. 2015; 4:791–9.
25. Ricarte-Filho JC, Ryder M, Chitale DA, Rivera M, Heguy A, Ladanyi M, et al. Mutational profile of advanced primary and metastatic radioactive iodine-refractory thyroid cancers reveals distinct pathogenetic roles for BRAF, PIK3CA, and AKT1. Cancer Res. 2009; 69:4885–93.

26. Pennelli G, Vianello F, Barollo S, Pezzani R, Merante Boschin I, Pelizzo MR, et al. BRAF(K601E) mutation in a patient with a follicular thyroid carcinoma. Thyroid. 2011; 21:1393–6.

28. Zou M, Baitei EY, Alzahrani AS, BinHumaid FS, Alkhafaji D, Al-Rijjal RA, et al. Concomitant RAS, RET/PTC, or BRAF mutations in advanced stage of papillary thyroid carcinoma. Thyroid. 2014; 24:1256–66.
29. Guerra A, Zeppa P, Bifulco M, Vitale M. Concomitant BRAF(V600E) mutation and RET/PTC rearrangement is a frequent occurrence in papillary thyroid carcinoma. Thyroid. 2014; 24:254–9.
Fig. 1.
Kaplan-Meier curves of relapse-free survival according to tumor stage and risk stratification. (A) Relapse-free survival of papillary thyroid carcinoma (PTC) patients according to American Joint Committee on Caner/Tumor-Nodes-Metastasis (AJCC/TNM) stage. (B) Relapse-free survival of PTC patients according to American Thyroid Association risk stratification.
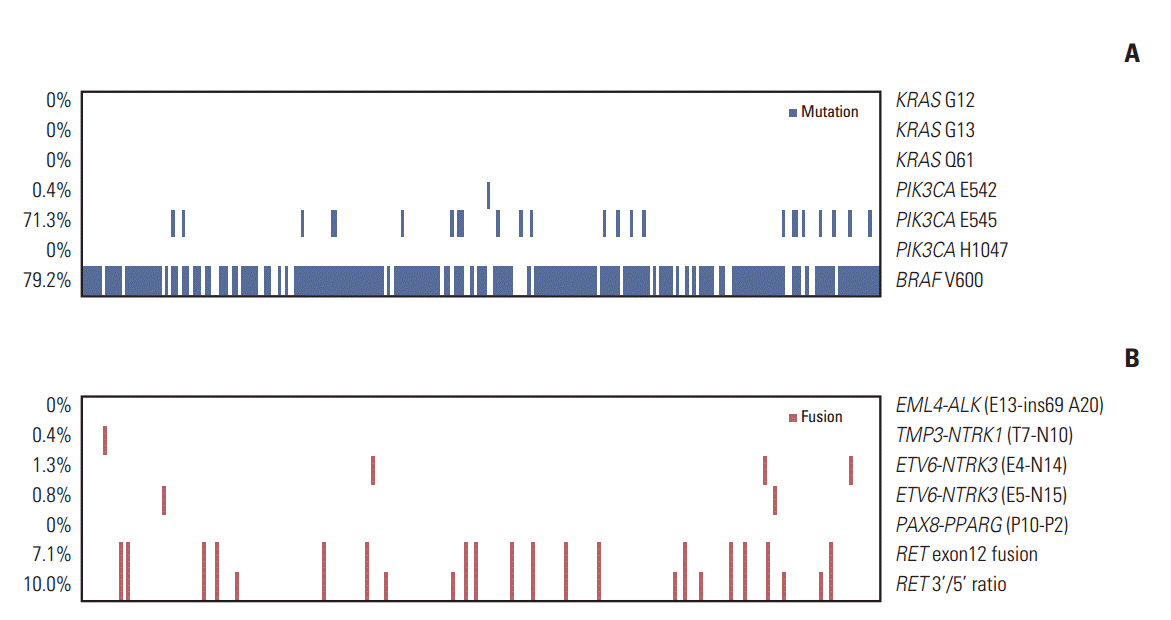
Fig. 2.
Mutations and fusions in high-risk papillary thyroid carcinoma. Heatmap of mutations (A) and fusions (B) found in 240 papillary thyroid carcinoma samples. The percentage of mutation and fusion incidence is noted at the left. The horizontal axis represents the complete dataset of patients. AJCC, American Joint Committee on Cancer; ATA, American Thyroid Association.
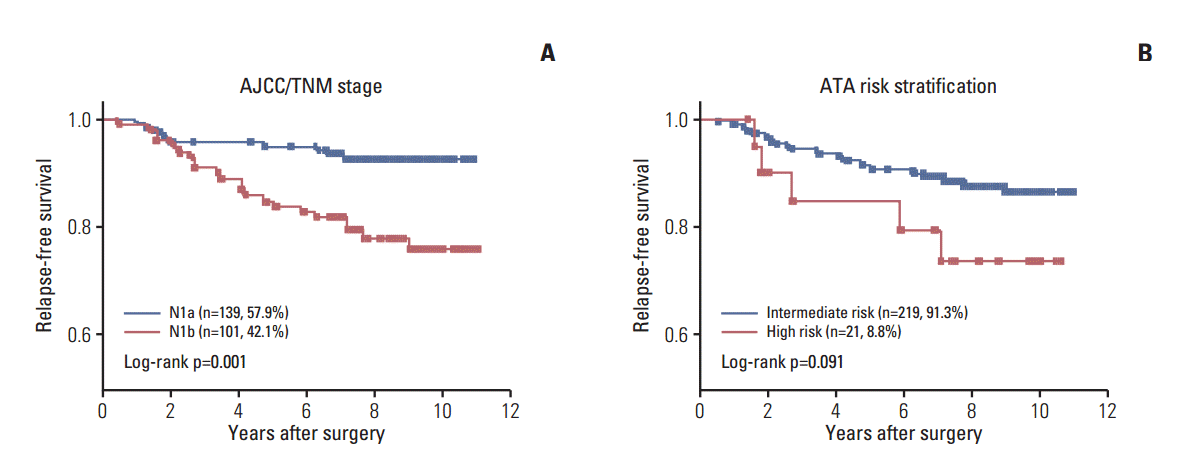
Fig. 3.
Impact of genetic alteration pattern on survival. (A) Relapse-free survival. (B) Overall survival. A Kaplan-Meier curve is presented according to the presence or absence of genetic alterations. Statistical analysis revealed no difference in recurrence-free probability among patients with no genetic alteration, one genetic alteration, two genetic alterations, or three genetic alterations.
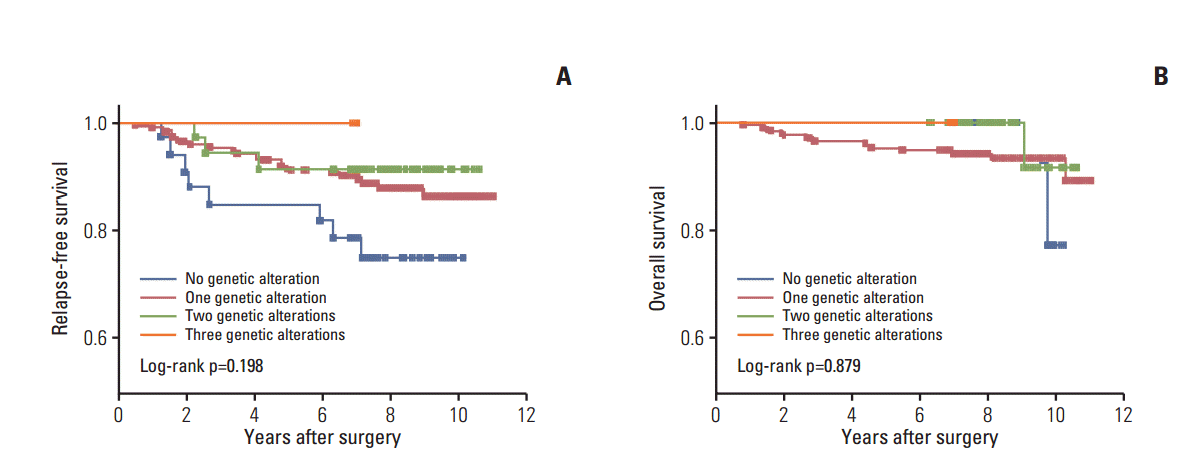
Table 1.
Baseline characteristics of the patients and tumor tissues
Table 2.
Concomitant existence of genetic alterations
Table 3.
Relationships of RET fusion gene arrangement with variables
| Variable |
RET fusion |
BRAF mutation |
||||
|---|---|---|---|---|---|---|
| Negative (n=216) | Positive (n=24) | p-value | Negative (n=50) | Positive (n=190) | p-value | |
| Age (yr) | ||||||
| < 45 | 94 (43.5) | 16 (66.7) | 0.031* | 28 (56.0) | 82 (43.2) | 0.105 |
| ≥ 45 | 122 (56.5) | 8 (33.3) | 22 (44.0) | 108 (56.8) | ||
| Sex | ||||||
| Male | 57 (26.4) | 8 (33.3) | 0.468 | 13 (36.0) | 52 (27.4) | 0.846 |
| Female | 159 (73.6) | 16 (66.7) | 37 (74.0) | 138 (72.6) | ||
| N stage | ||||||
| N1a | 132 (61.1) | 7 (29.2) | 0.003* | 26 (52.0) | 113 (59.5) | 0.341 |
| N1b | 84 (38.9) | 17 (70.8) | 24 (48.0) | 77 (40.5) | ||
| RET | ||||||
| Fusion (+) | - | - | - | 11 (22.0) | 13 (6.8) | 0.001* |
| Fusion (–) | - | - | - | 39 (78.0) | 177 (93.2) | |
| BRAF | ||||||
| Mutation (+) | 177 (81.9) | 13 (54.2) | 0.001* | - | - | - |
| Mutation (–) | 39 (18.1) | 11 (45.8) | - | - | - | |
| PIK3CA | ||||||
| Mutation (+) | 152 (70.4) | 19 (79.2) | 0.366 | 33 (66.0) | 138 (72.6) | 0.357 |
| Mutation (–) | 64 (29.6) | 5 (20.8) | 17 (34.0) | 52 (27.4) | ||
| NTKR1/3 | ||||||
| Fusion (+) | 6 (2.8) | 0 | > 0.990 | 2 (4.0) | 4 (2.1) | 0.607 |
| Fusion (–) | 210 (97.2) | 24 (100) | 48 (96.0) | 186 (97.9) | ||




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download