Abstract
Purpose
The biological function of long non-coding RNAs (lncRNAs) is only partially understood; therefore, in this study, we investigated the expression of the novel HOXA11 antisense (HOXA11as) lncRNA and its oncogenic role in serous ovarian cancer (SOC).
Materials and Methods
HOXA11as expression was examined in 129 SOC tissue samples by real time reverse transcription polymerase chain reaction. Clinicopathological factors and patient survival were compared between the high (n=27) and low HOXA11as expression group (n=102). To investigate the role of HOXA11as in cell proliferation, invasion, and migration, HOXA11as expression in ovarian cancer cells was knocked down using RNA interference.
Results
HOXA11as expression in cancer tissue was 77-fold higher than that of noncancerous tissue (p < 0.05). Higher HOXA11as expression was significantly correlated with histological grade (p=0.017) and preoperative cancer antigen 125 (p=0.048). HOXA11as overexpression in SOC cells led to increased cell proliferation, invasion, and migration. Moreover, HOXA11as was associated with the expression of genes involved in cell invasion, migration, and epithelial-mesenchymal transition (EMT), including vascular endothelial growth factor, matrix metalloproteinase 9 (MMP-9), B-catenin, E-cadherin, Snail, Twist, and vimentin. Multivariate analysis revealed that HOXA11as was a prognostic factor of progressive disease and mortality (hazard ratio [HR], 1.730; p=0.043 and HR, 2.170; p=0.033, respectively). Progression-free and overall survival were significantly shorter in patients with high HOXA11as expression.
Epithelial ovarian cancer has the highest mortality rate among cancers of the reproductive organs and is the fifth leading cause of female cancer death in the United States [1]. Despite advances in surgery and chemotherapeutic agents, the prognosis for ovarian cancer is poor, with a 5-year survival rate of less than 30% [2]. Serous ovarian cancer (SOC), which often presents at advanced stages and is characterized by metastases in the pelvic and abdominal cavity, is the most common histological type. The poor prognosis of SOC has been correlated with tumor metastasis and recurrence; therefore, it is crucial to understand the underlying molecular mechanisms involved in ovarian carcinogenesis and progression.
Long non-coding RNAs (lncRNAs) comprise a heterogenous group of genomic transcripts longer than 200 nucleotides that have no protein coding functions [3]. Unlike short non-coding RNAs such as microRNAs, the functional role of lncRNAs have been underestimated since they were initially regarded as transcriptional noise in the genome [4]; however, recent data have demonstrated their importance in normal physiology, as well as in the modulation of various biological processes such as cell proliferation, apoptosis, invasion, and reprogramming of stem cell pluripotency [4]. Furthermore, emerging evidence indicates that lncRNA expression is altered in diverse human cancers, and that its expression pattern may be associated with cancer progression and metastasis [5-7].
Members of the homeobox (HOX) family of genes are known to contain transcription factors that contribute to embryogenesis and carcinogenesis [8]. Several studies have shown dysregulated HOX gene expression in breast, lung, prostate, and colon cancer [9-13]. In humans, HOX genes are located in four different chromosomes, organized into four clusters (A, B, C, and D) [14]. During development of the female reproductive tract, HOXA11 is expressed in the cervix and lower uterine segment; however, its inappropriate expression is believed to lead to epithelial ovarian neoplasia since it promotes aberrant epithelial differentiation [15,16]. Similarly, the HOXA cluster of protein-coding genes contributes to ovarian embryogenesis and carcinogenesis. This study focused on the ‘antisense’ strand of the HOXA gene cluster, which contains non-coding RNA genes. The 5' region of the HOXA locus includes three protein-coding genes (HOXA9, HOXA10, and HOXA11) and three lncRNAs, HOTTIP, HOXA10as (antisense), and HOXA11as. Since little is known about the function of locally residing lncRNAs, this study was conducted to investigate the role of HOXA11as in carcinogenesis.
In this study, the expression of HOXA11as in SOC was examined and its clinical significance and correlation to disease prognosis were analyzed. Functional assays were also conducted to explore the impact of HOXA11as on cancer cell invasion and migration in vitro. Finally, since epithelial-mesenchymal transition (EMT) is regarded as one of the major mechanisms inducing cancer metastasis, we investigated whether HOXA11as was involved in EMT and metastasis in SOC.
A total of 129 SOC tissue samples were obtained from patients who underwent surgery at the Department of Obstetrics and Gynecology, Severance Hospital between April 2003 and December 2013. Patients with borderline ovarian tumor, concomitant gynecological or other primary cancer, as well as those who had received preoperative chemotherapy were excluded. Table 1 summarizes the clinical information describing the patients. The median duration of follow-up was 39 months (range, 2 to 116 months) for survivors. Progression-free survival (PFS) was defined as the interval between the date of surgery and the date of progression confirmed by imaging studies. Overall survival (OS) was defined as the date of surgery until the date of death. The control group consisted of 38 normal ovarian epithelial tissue samples obtained from patients that underwent simple hysterectomy or oophorectomy for benign uterine conditions.
All samples were immediately frozen in liquid nitrogen and stored at –80°C until RNA extraction. This research was approved by the Institutional Review Board of Severance Hospital, Yonsei University College of Medicine (No. 4-2009-0738), and written informed consent was obtained from study participants.
The human epithelial ovarian cancer cell lines OVCAR3 and SKOV3 were obtained from the Korean Cell Line Bank (KCLB, Seoul, Korea) and the A2780 cell line was purchased from the European Collection of Cell Cultures (ECACC, Sigma-Aldrich, St. Louis, MO). Ovarian cancer cell lines OVCA433, OVCA429, and TOV112D were provided by the Korea Gynecologic Cancer Bank through the Bio & Medical Technology Development Program of the Minister of Science, Information and Communication Technology and Future Planning (MSIP), Korea. Normal human ovarian surface epithelial (HOSE) cells were purchased from ScienCell Research Laboratories (San Diego, CA). OVCAR3, SKOV3, and A2780 cells were cultured in RPMI-1640 medium (Gibco-BRL, Gaithersburg, MD), while OVCA433, OVCA429, and TOV112D cells were cultured in Dulbecco's modified Eagle medium and the HOSE cell line was cultured in ovarian epithelial cell medium (OEpiCM, ScienCell). All culture media were supplemented with 10% (vol/vol) fetal bovine serum and 1% penicillin/streptomycin, and cell lines were maintained at 37°C in a humidified atmosphere of 5% CO2 and 95% air. Culture medium was replaced with fresh medium every 2-3 days and cells were used between passages 5 and 10.
Total RNA was extracted from tissues or cultured cells using TRIzol reagent (Invitrogen, Carlsbad, CA). For quantitative real time polymerase chain reaction (qRT-PCR), total RNA was reverse transcribed to cDNA using a Reverse Transcription Reagent Kit (Invitrogen) according to the manufacturer’s protocols. Real-time PCR analyses were conducted using a SYBR Green Real-time PCR Kit (TOYOBO Co. Ltd., Osaka, Japan). Conditions for the amplification of HOXA11as lncRNA were as follows: initial denaturation at 95°C for 3 minutes, followed by 40 cycles of denaturation at 95°C for 15 seconds, annealing at 60°C for 60 seconds, and elongation at 72°C for 60 seconds and then final elongation at 72°C for 5 minutes. qRT-PCR was performed using an ABI StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA). The results were normalized to the expression of U6. The polymerase chain reaction (PCR) primer sequences used for analyses were as follows: HOXA11as, 5′-GAGTTTGAAGCCGTGGATGT-3′ (sense) and 5′-AGATGAGGGGAGAGGTGGAT-3′ (antisense); E-cadherin, 5′-ATTCTGATTCTGCTGCTCTTG-3′ (sense) and 5′- AGTAGTCATAGTCCTGGTCCT-3′ (antisense); N-cadherin, 5′-CCCAAGACAAGAGACCCAG-3′ (sense) and 5′-GCCACTGTGCTTACTGAATTG-3′ (antisense); β-catenin, 5′-TGCAGTTCG CCTTCACTATG-3′ (sense) and 5′-ACTAGTCGTGGAATGGCACC-3′ (antisense); vimentin, 5′-TGGATTCACTCCCTCTGGTT-3′ (sense) and 5′-GGTCATCGTGATGCTGAGAA-3′ (antisense); Snail, 5′-GAGGCGGTGGCAGACTAG-3′ (sense) and 5′-GACACATCGGTCAGACCAG-3′ (antisense); Twist, 5′-CGGGAGTCCGCAGTCTTA-3′ (sense) and 5′-TGAATCTTGCTCAGCTTGTC-3′ (antisense); U6, 5′-CTCGCTTCGGCAGCACA-3′ (sense) and 5′-AACGCTTCAGGAATTTGCG T-3′ (antisense). The relative change in expression of mRNA was calculated by the 2–ΔΔCT method. All qRT-PCR experiments were replicated at least three times.
Homeobox A11 antisense lncRNA small interfering RNA (siRNA) (siHOXA11as) and negative control siRNA (siNC) were purchased from Genolution (Genolution Pharmaceuticals Inc., Seoul, Korea). Cells (5×104 cells/well) were seeded into 6-well plates and transfected with 10 nM siRNA in OptiMEM I (Invitrogen) using the Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions. These siRNA-transfected cells were used for in vitro assays 48-hour post-transfection. The target sequence for siHOXA11as was as follows: siRNA, 5′-CGGAAUAUCGGAAUAAAGUUU-3′. All experiments were repeated at least three times.
Full-length human HOXA11as transcript cDNA was amplified by PCR and inserted into the pLenti6/V5-D-TOPO vector according to the ViraPower Lentiviral Expression Systems (Invitrogen) protocol. Briefly, the plasmid was transfected into 293FT cells for packaging, after which the lentivirus was used to infect the desired cell lines. The selection of HOXA11as stably transfected cells was performed in medium containing 10 μg/mL blasticidin (Invitrogen).
Cell proliferation was measured by a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Sigma). Briefly, cells (1×104 cells/well) were seeded into 96-well flat-bottomed plates in 100 μL of complete medium. The cells were then incubated overnight to allow cell attachment and recovery, after which they were transfected with siNC or siHOXA11as for 24, 48, 72, or 96 hours. MTT solution (10 μL) was then added to each well, after which the cells were incubated for an additional 2 hours. Absorbance was measured at 570 nm using a microplate reader. Three independent experiments were performed in triplicate.
Cell migration was assessed by monolayer wound healing assay. Briefly, cells were seeded into 6-well culture plates with serum-containing medium and allowed to grow to 90% confluency. The serum-containing medium was then removed, after which cells were serum starved for 24 hours. When the cell confluence reached nearly 100%, an artificial homogenous wound was created by scratching the monolayer with a sterile 200 μL pipette tip. After the cells were washed with serum-free medium, images of cells migrating into the wound were captured at 0, 24, and 48/60 hours using a microscope. Each experiment was repeated three times.
Matrigel invasion assay was performed using the BD Biocoat Matrigel Invasion Chamber (pore size, 8 μm; 24-well; BD Biosciences, Bedford, MA) according to the manufacturer's instructions. Briefly, overexpression cells or siHOXA11astransfected cells and siNC-transfected cells (5×104 cells/mL) were plated in the upper chamber in serum-free medium, and complete medium was added to the bottom chamber. After 24 hours of incubation, cells that had invaded through the membrane were stained using a Differential Quik Stain Kit (Diff Quik, Sysmex, Kobe, Japan), then counted using a light Microscopy Axio Imager M2 (Carl Zeiss, Thornwood, NY; magnification ×200). The assay was performed in triplicate.
Proteins were extracted with RIPA buffer (Thermo Fisher Scientific, Inc., Waltham, MA) and their concentrations were determined using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). Next, samples were boiled for 5 minutes, subjected to 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and transferred electrophoretically to polyvinylidene difluoride membranes (Millipore, Billerica, MA). The membranes were subsequently blocked with 5% non-fat dried milk in 1× Tris-buffered saline containing 0.1% Tween 20 (TBST; pH 7.6) at room temperature for 1 hour, then incubated with primary antibody at 4°C overnight. Blocked membranes were then incubated with primary antibodies (E-cadherin, N-cadherin, or β-catenin rabbit polyclonal antibodies at final concentrations of 1:1,000, Cell Signaling, Beverly, MA; vimentin, Snail, or β-actin mouse polyclonal antibodies at a dilution of 1:1,000, Sigma). Primary antibodies against each protein were detected by horseradish peroxidase-conjugated secondary antibody (1:2,000). Bands were visualized using a SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific), and band intensities were quantified using the Luminescent Image Analyzer (LAS-4000 mini, Fujifilm, Uppsala, Sweden).
Statistical analysis was performed with SPSS ver. 19 for Windows (SPSS Inc., Chicago, IL). The Kolmogorov-Smirnov test was used to verify standard normal distributional assumptions. A student’s t test and the Mann-Whitney U test were used for parametric and non-parametric variables, respectively. Differences between proportions were compared using Fisher exact test or a chi-square test. PFS and OS were calculated by the Kaplan-Meier method, while the log-rank test was used to compare survival distribution. Multivariate analysis using the Cox regression model adjusted for known prognostic covariates (age, stage, tumor grade, lymph node metastasis, and residual tumor) was conducted. A p < 0.05 was regarded as statistically significant.
The expression level of HOXA11as was measured in 129 SOC tissues and 38 corresponding normal ovarian tissues by qRT-PCR and normalized to U6. HOXA11as expression in cancer tissue was more than 77-fold higher than that of non-cancerous tissue (p < 0.05) (Fig. 1A). One hundred twenty nine SOC patients were divided into a high (n=27) and low expression group (n=102) based on a HOXA11as/U6 ratio of 60 in cancerous tissue (Fig. 1B).
Clinicopathological data such as age, stage, histologic grade, extent of residual disease, lymph node metastasis, and preoperative cancer antigen 125 (CA125) level were compared between the high and low HOXA11as expression groups (Table 1). High grade histology and higher level of CA125 were more frequently found in the high HOXA11as expression group (p < 0.05).
To investigate the role of HOXA11as in SOC cells, several cell lines were examined for HOXA11as expression. As shown in Fig. 2A, OVCA429, OVCAR3, and SKOV3 expressed higher levels of HOXA11as than other cell lines and the control (HOSE). The knockdown efficiency of the HOXA11as-specific siRNAs (siHOXA11as) was analyzed by qRT-PCR method, which revealed that siHOXA11as had higher silencing efficiency compared to control (Fig. 2B). The proliferation of siHOXA11as-transfected ovarian cancer cells and siNC was measured by MTT assay. Knockdown of HOXA11as inhibited cell proliferation by 40% and 50% at 96 hours post-transfection in OVCA429 and SKOV3 cell lines, respectively, relative to control cells (Fig. 2C and D).
The effects of HOXA11as on the invasive and migratory behavior of SOC cells were assessed by Matrigel invasion and wound healing assays. Wound healing assays showed larger width of wound in siHOXA11as-transfected OVCA 429 and SKOV3 cells than siNC-transfected cells, which demonstrated decreased migration of SOC cells via down-regulation of HOXA11as (Fig. 3A and B). According to the Matrigel invasion assay, the knockdown of HOXA11as significantly reduced the invasive cell numbers by more than 80% (Fig. 3C). The same results were obtained from migration and invasion assays after enforcing HOXA11as expression in OVCA429 cells (Fig. 3D). The invasive capacity of OVCA429 cells increased after 48 hours upon overexpression of HOXA11as (Fig. 3E and F). Taken together, these results indicate that HOXA11as promotes SOC cell invasion and migration in vitro.
The expression of proteins related to tumor progression and metastasis were determined in SOC cells to evaluate the possible mechanism of HOXA11as in cancer cell invasion and migration. As shown in Fig. 4A, knockdown of HOXA11as inhibited the level of matrix metalloproteinase (MMP) 2, MMP-9, and vascular endothelial growth factor (VEGF) expression in OVCA429 cells. The expression of these proteins was further validated by western blot analyses, which showed significantly lower expression in siHOXA11-as transfected cells than siNC-transfected cells (Fig. 4B). Taken together, these findings indicate that HOXA11as regulates SOC cell invasion and migration through the regulation of MMP and VEGF.
To further investigate the molecular mechanisms involved in metastatic features of HOXA11as, the EMT marker gene levels were examined using quantitative PCR and western blot assays. Epithelial marker E-cadherin was significantly upregulated and mesenchymal marker N-cadherin, β-catenin, and vimentin were downregulated in siHOXA11as cells. The impact of HOXA11as knockdown on the expression of Twist and Snail, which are known to modulate EMT, was also assessed. siHOXA11as-transfected cells showed lower expression of Twist and Snail than siNC-transfected cells (Fig. 5).
The median duration of follow up was 39 months (range, 2 to 116 months). Overall, 45 patients (34.9%) died from the disease, while 91 (70.5%) had disease recurrence. Survival analysis was performed to measure the PFS and OS of patients with SOC with different HOXA11as expression. As shown in Fig. 6A, the 10-year PFS of patients who had high HOXA11as cancer tissue expression was unfavorable relative to those with low HOXA11as expression (median PFS, 12 months vs. 24 months; p=0.013). Similarly, worse OS outcomes were shown in the high HOXA11as expression group than the low expression group (median OS, 53 months vs. 77 months; p=0.045) (Fig. 6B). Furthermore, the receiver operating characteristic curve analysis showed that the HOXA11as level was useful to predict survival of SOC patients (area under the curve, 0.731; 95% confidence interval [CI], 0.646 to 0.817) (Fig. 6C). Multivariate Cox regression analysis for survival revealed that high HOXA11as expression in tumor tissues was an independent predictor of poor PFS (hazard ratio [HR], 1.730; 95% CI, 1.015 to 2.948; p=0.043) and OS (HR, 2.170; 95% CI, 1.062 to 4.431; p=0.033), regardless of age, stage, grade, residual tumor size and lymph node metastasis (Table 2). Upon multivariate analysis for overall survival, age (HR, 1.044; 95% CI, 1.008 to 1.081; p=0.015) and residual tumor size of > 1.0 cm (HR, 2.514; 95% CI, 1.223 to 5.167; p=0.012) were associated with mortality, but not with recurrence.
In this study, HOXA11as expression levels in SOC were higher than those of noncancerous tissues, and increased HOXA11as expression was correlated with poor patient survival. Moreover, knockdown of HOXA11as expression led to reduced cell proliferation, invasion and migration in SOC cells. The metastatic effects of HOXA11as were related to the regulation of genes involved in cell invasion, migration, and EMT, including VEGF, MMP-9, B-catenin, E-cadherin, Snail, Twist, and vimentin. These findings highlighted the clinical relevance of HOXA11as to predicting adverse prognosis of SOC and suggested its potential in promoting tumor aggressiveness via the regulation of VEGF and EMT-related mechanisms.
In recent years, accumulating evidence has shown that lncRNAs may play a critical role in cellular biology and human diseases. Several lncRNAs have been identified in gynecological cancer, including HOTAIR, MALAT-1, H19, and LSINCT5 [17]. One of the most popular oncogenic lncRNA studied is HOX transcript antisense intergenic RNA (HOTAIR), which our group recently identified as playing a role in cell proliferation and invasion in cervical cancer cell lines [18]. However, no studies have been published regarding the clinical/prognostic significance of the novel lncRNA HOXA11as in SOC.
The role of HOXA11as, a novel lncRNA in epithelial ovarian cancer has been underexplored. In this study, HOXA11as expression was shown to be higher in cancer tissues, and knockdown of HOXA11as inhibited cell proliferation in various ovarian cancer cell lines. Furthermore, overexpression of HOXA11as in OVCA429 cells enhanced the invasive capacity at 48 hours, suggesting that HOXA11as contributes to the invasive and migratory phenotype of ovarian carcinoma cells. LncRNAs that are overexpressed in ovarian tumors similar to HOXA11as include AB073614, HOST2, LSINCT5, HOTAIR, and H19 [19-22]. In vitro and in vivo observations of these lncRNAs have shown decreased cell growth, migration, and apoptosis when downregulated as in this study. Among these, HOST2 has been proposed to modulate the availability of tumor suppressor that subsequently suppresses the expression of targets that regulate cell growth and motility at the post-transcriptional level [20]. These observations are relevant to HOXA11as because its upregulation is similarly associated with cancer cell growth and migration. To determine if HOXA11as promotes SOC metastasis by regulating genes that encode certain metastasis-related proteins, the expression of extracellular matrix degrading protease MMPs and VEGF were analyzed. MMPs degrade basement membrane collagen, which promotes tumor cell invasion and metastasis. These compounds are also known to decrease survival in several types of cancers [23]. VEGF, together with MMPs, is known for its role in tumor angiogenesis and essential for cell motility and metastasis [24]. As expected, our results demonstrated significantly decreased expression of MMP-2, MMP-9, and VEGF with downregulation of HOXA11as. These findings indicate that HOXA11as plays a role in the aggressive nature of ovarian cancer cells through upregulation of the possible down-stream targets, MMPs and VEGF.
Since the molecular mechanisms of lncRNAs related to tumor progression and metastasis are not fully understood, a question was raised as to whether HOXA11as promotes SOC metastasis by regulating gene expression that encodes metastasis-related proteins. One of the possible mechanisms is thought to be EMT, which involves epithelial cells with mesenchymal properties such as reduced cell-cell junction and increased motility. These properties of EMT have been reported to contribute to cellular proliferation, invasion and migration in various malignancies [25,26]. The knockdown of lncRNA HOTAIR was recently shown to be associated with reversal of the EMT process in gastric cancer cells [27], and lncRNA MALAT1 was reported to function as an inducer of EMT in breast cancer by activating the phosphoinositide 3-kinase–AKT pathway [28]. Similarly, genes related to EMT (E-cadherin, N-cadherin, B-catenin, Snail, and vimentin) were dysregulated by HOXA11as knockdown in serous type OVCA429 cells in the present study. Moreover, E-cadherin was markedly upregulated and the mesenchymal markers N-cadherin, β-catenin and vimentin were downregulated in siHOXA11as cells. Furthermore, siHOXA11as-transfected cells showed lower expression levels of Twist and Snail than siNC-transfected cells, indicating HOXA11as-mediated regulation of EMT modulators. Taken together, dysregulated expression of EMT-related genes appears to take part in HOXA11as-related SOC cell invasion and migration.
The results of this study suggest the possible relationship of HOXA11as with the EMT and contribute to our understanding of the role of lncRNAs. Additionally, this is the first investigation to demonstrate the correlation between the novel lncRNA HOXA11as and SOC cell metastasis in relation to EMT. It should be noted that this study was limited by its retrospective nature; however, a large number of cases were examined along with long term survival data. Many studies have shown that lncRNA acts as an unfavorable prognostic factor in various human cancers [17,18,29]; thus, the clinical and prognostic significance of HOXA11as were identified in this study. Moreover, only serous type histology was investigated to overcome the heterogeneity of the study population. Further investigations are required to understand the role and molecular mechanisms of HOXA11as in other subtypes of ovarian cancer.
In summary, HOXA11as was overexpressed in patients with SOC, and its overexpression was correlated with poor prognosis. Furthermore, functional studies suggested that HOXA11as plays a critical role in controlling SOC cell proliferation and invasion via regulation of EMT-related genes. These findings contribute to a better understanding of dysregulated lncRNAs in cancer progression and may provide guidance for the development of lncRNA-based biomarkers and precision medicine approaches for treatment of SOC.
ACKNOWLEDGMENTS
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (NRF-2015R1A2A2A01008162; NRF-2015R1C1-A2A01053516).
References
3. Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014; 1839:1097–109.

4. Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009; 23:1494–504.

5. Chen T, Xie W, Xie L, Sun Y, Zhang Y, Shen Z, et al. Expression of long noncoding RNA lncRNA-n336928 is correlated with tumor stage and grade and overall survival in bladder cancer. Biochem Biophys Res Commun. 2015; 468:666–70.

6. Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013; 73:1180–9.

7. Hu L, Wu Y, Tan D, Meng H, Wang K, Bai Y, et al. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2015; 34:7.

8. Bhatlekar S, Fields JZ, Boman BM. HOX genes and their role in the development of human cancers. J Mol Med (Berl). 2014; 92:811–23.

9. Calvo R, West J, Franklin W, Erickson P, Bemis L, Li E, et al. Altered HOX and WNT7A expression in human lung cancer. Proc Natl Acad Sci U S A. 2000; 97:12776–81.

10. Cantile M, Cindolo L, Napodano G, Altieri V, Cillo C. Hyperexpression of locus C genes in the HOX network is strongly associated in vivo with human bladder transitional cell carcinomas. Oncogene. 2003; 22:6462–8.

11. Miller GJ, Miller HL, van Bokhoven A, Lambert JR, Werahera PN, Schirripa O, et al. Aberrant HOXC expression accompanies the malignant phenotype in human prostate. Cancer Res. 2003; 63:5879–88.
12. Raman V, Martensen SA, Reisman D, Evron E, Odenwald WF, Jaffee E, et al. Compromised HOXA5 function can limit p53 expression in human breast tumours. Nature. 2000; 405:974–8.

13. Vider BZ, Zimber A, Hirsch D, Estlein D, Chastre E, Prevot S, et al. Human colorectal carcinogenesis is associated with deregulation of homeobox gene expression. Biochem Biophys Res Commun. 1997; 232:742–8.

14. Garcia-Fernandez J. The genesis and evolution of homeobox gene clusters. Nat Rev Genet. 2005; 6:881–92.

15. Cheng W, Liu J, Yoshida H, Rosen D, Naora H. Lineage infidelity of epithelial ovarian cancers is controlled by HOX genes that specify regional identity in the reproductive tract. Nat Med. 2005; 11:531–7.

16. Kelly ZL, Michael A, Butler-Manuel S, Pandha HS, Morgan RG. HOX genes in ovarian cancer. J Ovarian Res. 2011; 4:16.

17. Zhao M, Qiu Y, Yang B, Sun L, Hei K, Du X, et al. Long noncoding RNAs involved in gynecological cancer. Int J Gynecol Cancer. 2014; 24:1140–5.

18. Kim HJ, Lee DW, Yim GW, Nam EJ, Kim S, Kim SW, et al. Long non-coding RNA HOTAIR is associated with human cervical cancer progression. Int J Oncol. 2015; 46:521–30.

19. Cheng Z, Guo J, Chen L, Luo N, Yang W, Qu X. A long noncoding RNA AB073614 promotes tumorigenesis and predicts poor prognosis in ovarian cancer. Oncotarget. 2015; 6:25381–9.

20. Gao Y, Meng H, Liu S, Hu J, Zhang Y, Jiao T, et al. LncRNA-HOST2 regulates cell biological behaviors in epithelial ovarian cancer through a mechanism involving microRNA let-7b. Hum Mol Genet. 2015; 24:841–52.

21. Silva JM, Boczek NJ, Berres MW, Ma X, Smith DI. LSINCT5 is over expressed in breast and ovarian cancer and affects cellular proliferation. RNA Biol. 2011; 8:496–505.

22. Yan L, Zhou J, Gao Y, Ghazal S, Lu L, Bellone S, et al. Regulation of tumor cell migration and invasion by the H19/let-7 axis is antagonized by metformin-induced DNA methylation. Oncogene. 2015; 34:3076–84.

23. Curran S, Murray GI. Matrix metalloproteinases: molecular aspects of their roles in tumour invasion and metastasis. Eur J Cancer. 2000; 36:1621–30.
24. Geng YJ, Xie SL, Li Q, Ma J, Wang GY. Large intervening noncoding RNA HOTAIR is associated with hepatocellular carcinoma progression. J Int Med Res. 2011; 39:2119–28.

25. Baum B, Settleman J, Quinlan MP. Transitions between epithelial and mesenchymal states in development and disease. Semin Cell Dev Biol. 2008; 19:294–308.

26. Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006; 7:131–42.

27. Xu ZY, Yu QM, Du YA, Yang LT, Dong RZ, Huang L, et al. Knockdown of long non-coding RNA HOTAIR suppresses tumor invasion and reverses epithelial-mesenchymal transition in gastric cancer. Int J Biol Sci. 2013; 9:587–97.

28. Xu S, Sui S, Zhang J, Bai N, Shi Q, Zhang G, et al. Downregulation of long noncoding RNA MALAT1 induces epithelialto-mesenchymal transition via the PI3K-AKT pathway in breast cancer. Int J Clin Exp Pathol. 2015; 8:4881–91.
29. Huang C, Yu W, Wang Q, Cui H, Wang Y, Zhang L, et al. Increased expression of the lncRNA PVT1 is associated with poor prognosis in pancreatic cancer patients. Minerva Med. 2015; 106:143–9.
Fig. 1.
HOXA11 antisense (HOXA11as) expression in human ovarian tumor tissue. (A) Relative expression of HOXA11as was significantly higher in serous ovarian cancer (SOC) tissues (n=129) than in noncancerous tissues (n=38). HOXA11as expression was determined using quantitative real time polymerase chain reaction with U6 as an internal control. (B) HOXA11as expression was classified into two groups according to the expression level of SOC tissues. Data are expressed as mean±standard deviation. *p < 0.05 vs. non-tumor control.
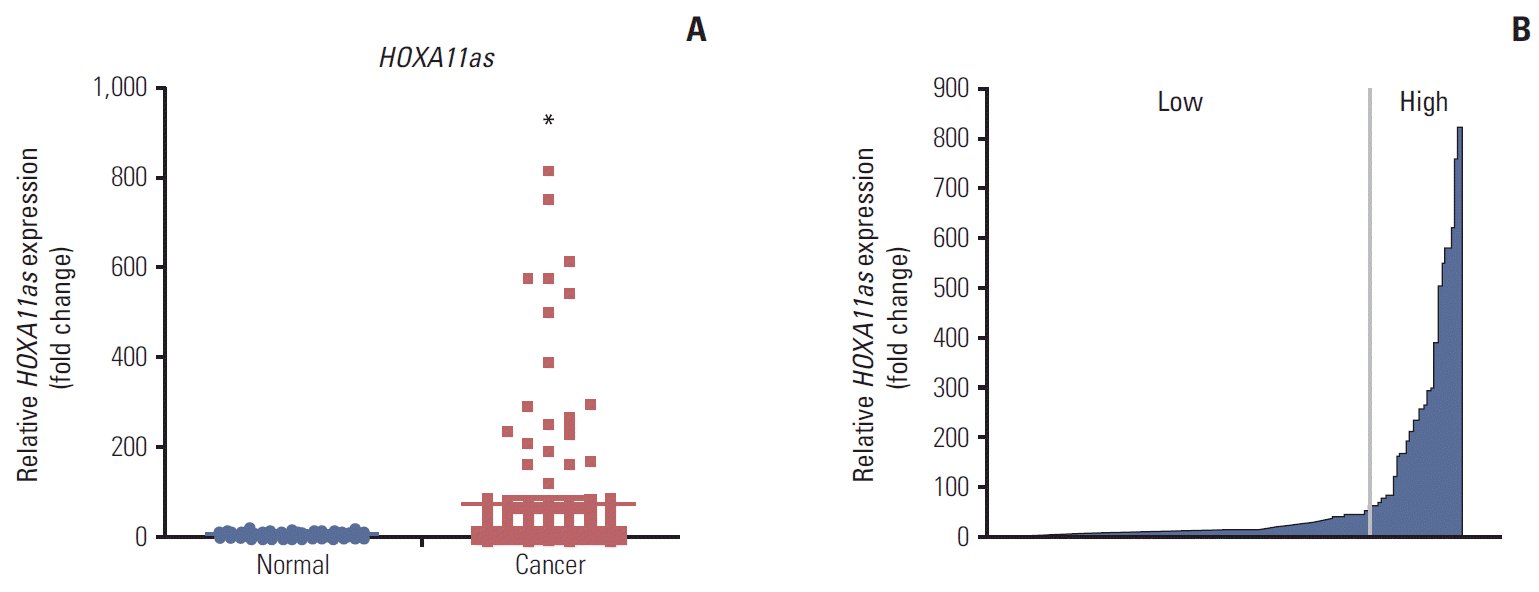
Fig. 2.
Knockdown of HOXA11 antisense (HOXA11as) inhibits serous ovarian cancer cell proliferation. (A) Expression of HOXA11as in human ovarian surface epithelial cell line (HOSE) and six ovarian cancer cell lines determined by quantitative real time polymerase chain reaction (qRT-PCR). (B) Knockdown efficiency was determined by qRT-PCR analysis in OVCA429 and SKOV3 cells. Cells were transfected with HOXA11as-specific siRNA (siHOXA11as) and negative control siRNA (siNC). (C, D) Knockdown of HOXA11as significantly reduced cell proliferation in OVCA429 and SKOV3 cells as determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Bars indicate mean±standard deviation of three independent experiments performed in triplicate. *p < 0.05 vs. siNC. siHOXA11as, HOXA11as-specific siRNA.
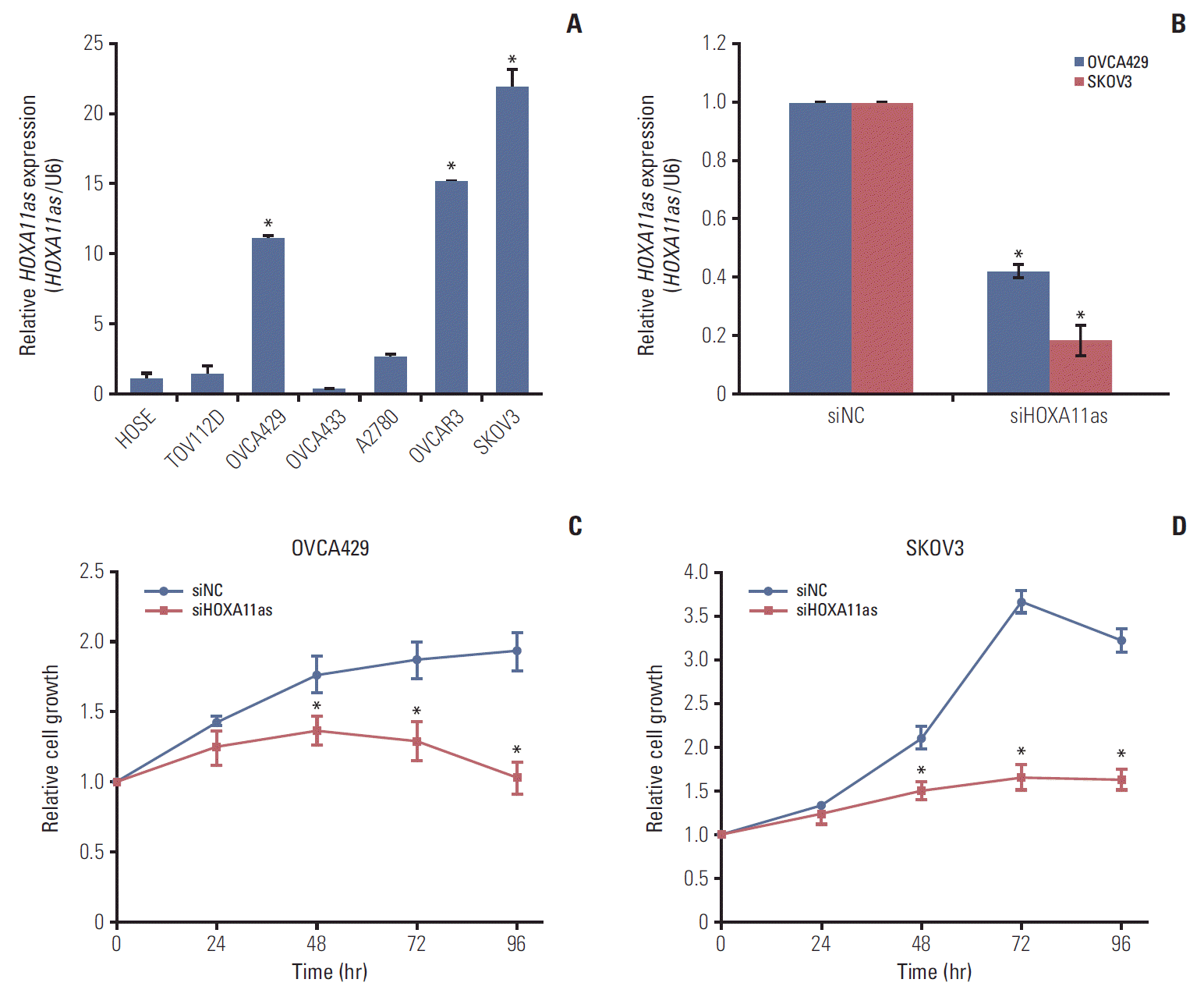
Fig. 3.
HOXA11 antisense (HOXA11as) promotes migration and invasion of ovarian cancer cells. (A, B) Wound healing assay was used to determine migration in HOXA11as-specific siRNA (siHOXA11as)-transfected OVCA429 and SKOV3 cells (×200). (C) Matrigel invasion assay was used to determine invasion after 24 hours in OVCA429 cells. (D) Overexpression of HOXA11as in OVCA429 cells analyzed by quantitative real time polymerase chain reaction. (E, F) Migration and invasion assay after overexpressing HOXA11as expression in OVCA429 cells. Overexpression of HOXA11as in OVCA429 cells increased the invasive capacity after 48 hours. Each assay was performed in triplicate. Data are mean±standard deviation. *p < 0.05 vs. siNC, vector.
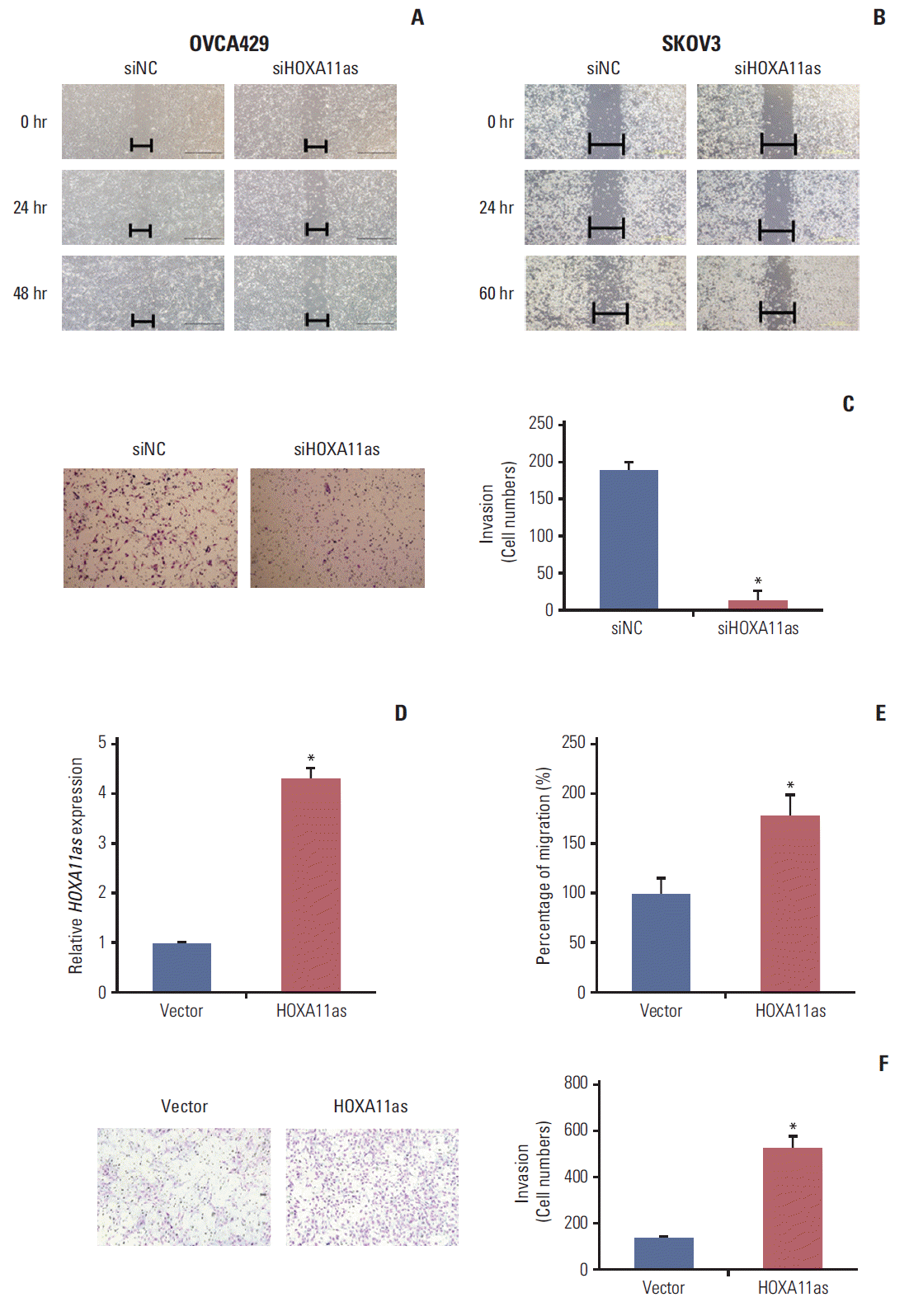
Fig. 4.
Knockdown of HOXA11 antisense (HOXA11as) inhibits matrix metalloproteinase (MMP) 2, MMP-9, and vascular endothelial growth factor (VEGF) expression in ovarian cancer cells. Protein lysates were obtained from HOXA11as-specific siRNA (siHOXA11as) and negative control siRNA (siNC)-transfected OVCA429 cells 48-hour post-transfection. MMP-2, MMP-9, and VEGF expression were analyzed by quantitative real time polymerase chain reaction (A) and western blotting (B). Each assay was performed in triplicate. Band intensities were quantitated, and MMP-2, MMP-9, and VEGF protein levels were normalized to that of β-actin. Each assay was performed in triplicate. Data are mean±standard deviation. *p < 0.05 vs. siNC.
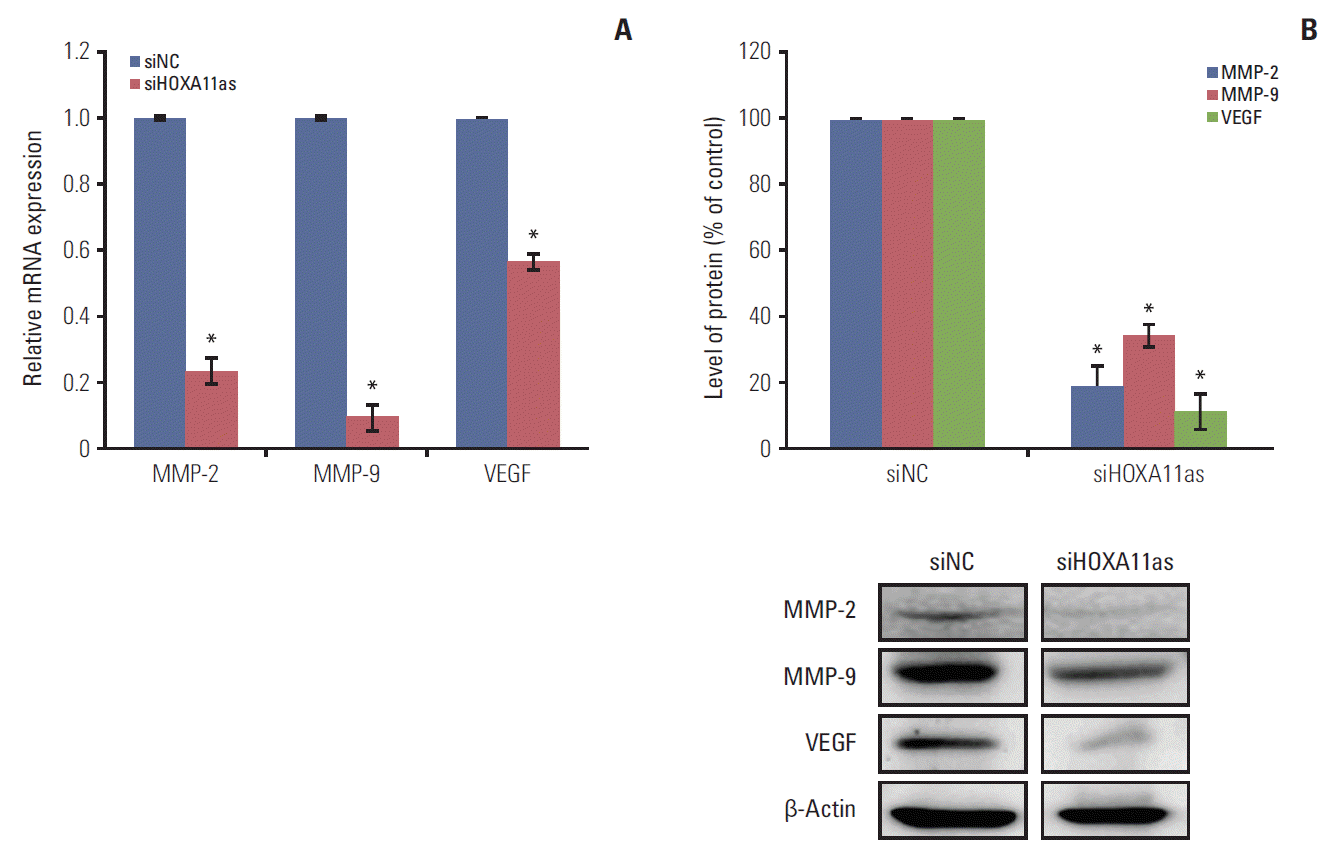
Fig. 5.
Expression of HOXA11 antisense (HOXA11as) knockdown on the epithelial-mesenchymal transition–related genes in OVCA429 cells. OVCA429 cells were transfected with HOXA11as-specific siRNA (siHOXA11as) and negative control siRNA (siNC) for 48 hours. E-cadherin, β-catenin, N-cadherin, vimentin, Snail, and Twist expression were analyzed by quantitative real time polymerase chain reaction (A) and western blotting (B). Each assay was performed in triplicate. Data are mean±standard deviation. *p < 0.05 vs. siNC.
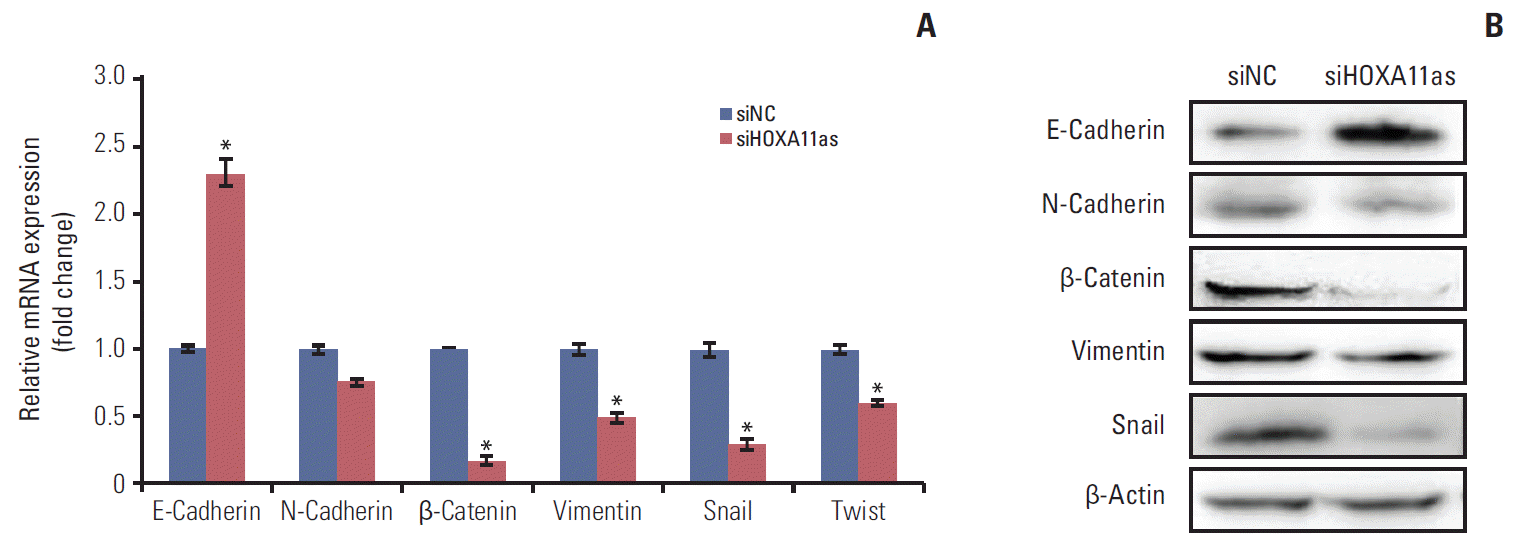
Fig. 6.
Correlation of HOXA11 antisense (HOXA11as) expression with patient survival. Kaplan-Meier curves for progression- free survival (A) and overall survival (B) in serous ovarian cancer patients with different expression levels of HOXA11as. (C) Receiver operating characteristic (ROC) curve for prognosis prediction of patients using HOXA11as level. The area under curve (AUC) is shown in plots.
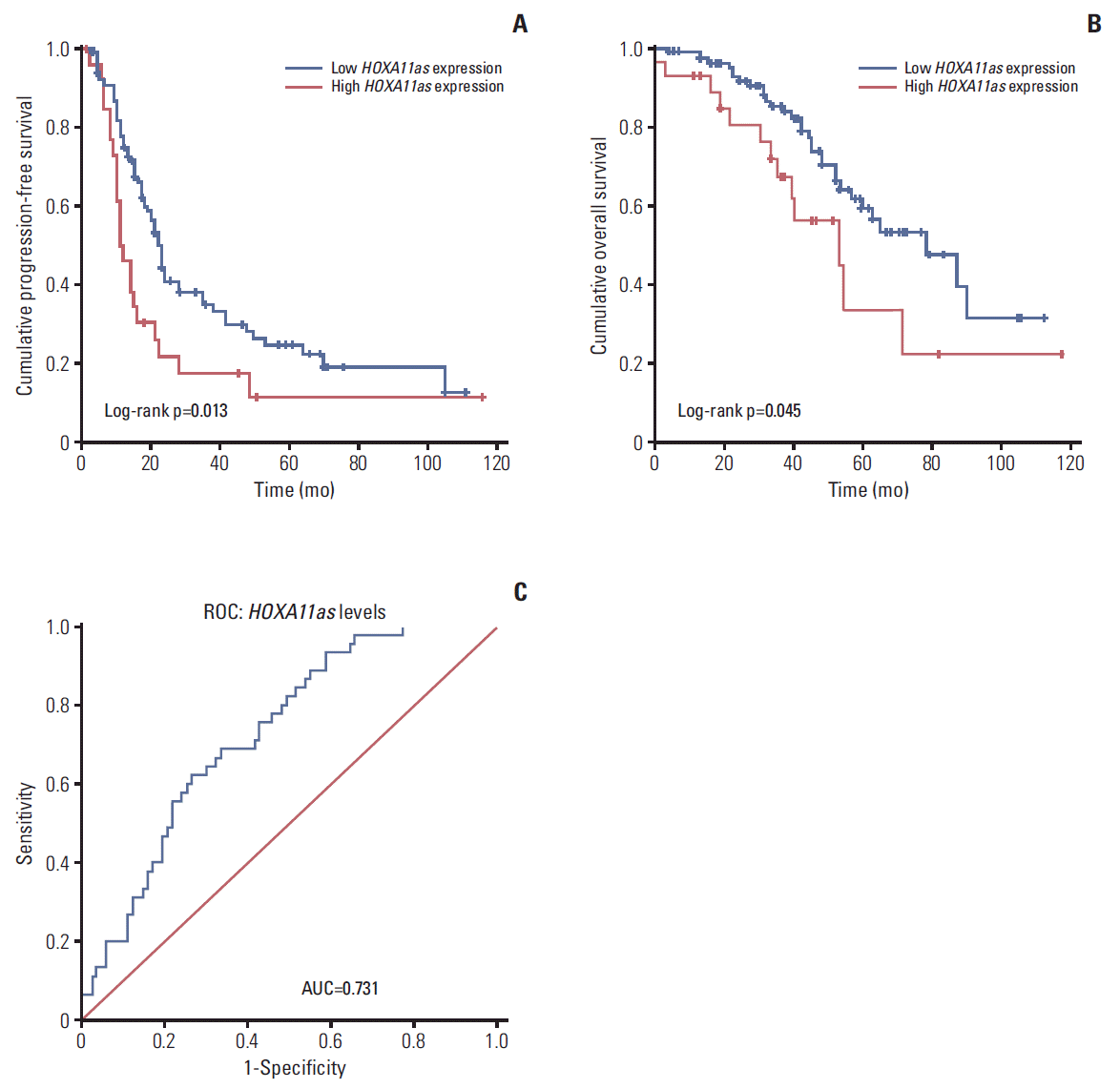
Table 1.
HOXA11as expression and clinicopathologic variables in serous ovarian cancer patients (n=129)
Table 2.
Multivariate Cox regression model analyses of factors associated with survival




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download