Introduction
Materials and Methods
1. Patients and samples
2. Gene expression microarray data analysis
Table 1.
3. qRT-PCR and analysis
4. Tissue microarray construction and immunohistochemical staining
5. Cell culture and chemotherapeutic agents
6. Establishment of stable cell line
7. Boyden chamber assay
8. Cell viability assay and clonogenicity
9. Immunoblotting
10. Statistical analysis for survival data
Results
1. Prognostic significance of ERO1L expression in gastric cancer patients
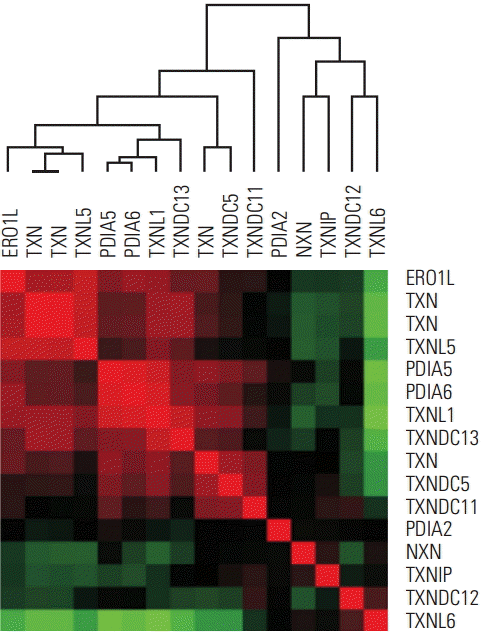 | Fig. 1.ERO1L and TXN family in-trans correlation in human gastric cancer. Data are given in NCBI’s GEO public database (microarray data accession number, GSE13861). The color red or green reflects relative high or low expression level, respectively. ERO1L, endoplasmic reticulum oxidoreduction 1-α; TXN, thioredoxin. |
 | Fig. 2.Two different conditions of gastric cancer cell lines, normoxia and hypoxia, show mRNA expression patterns. mRNA expression patterns of gastric cancer cells (AGS, NCI-N87, and SNU1) in normoxia and hypoxia are depicted in a heat map. The color red or green reflects relative high or low expression level, respectively. |
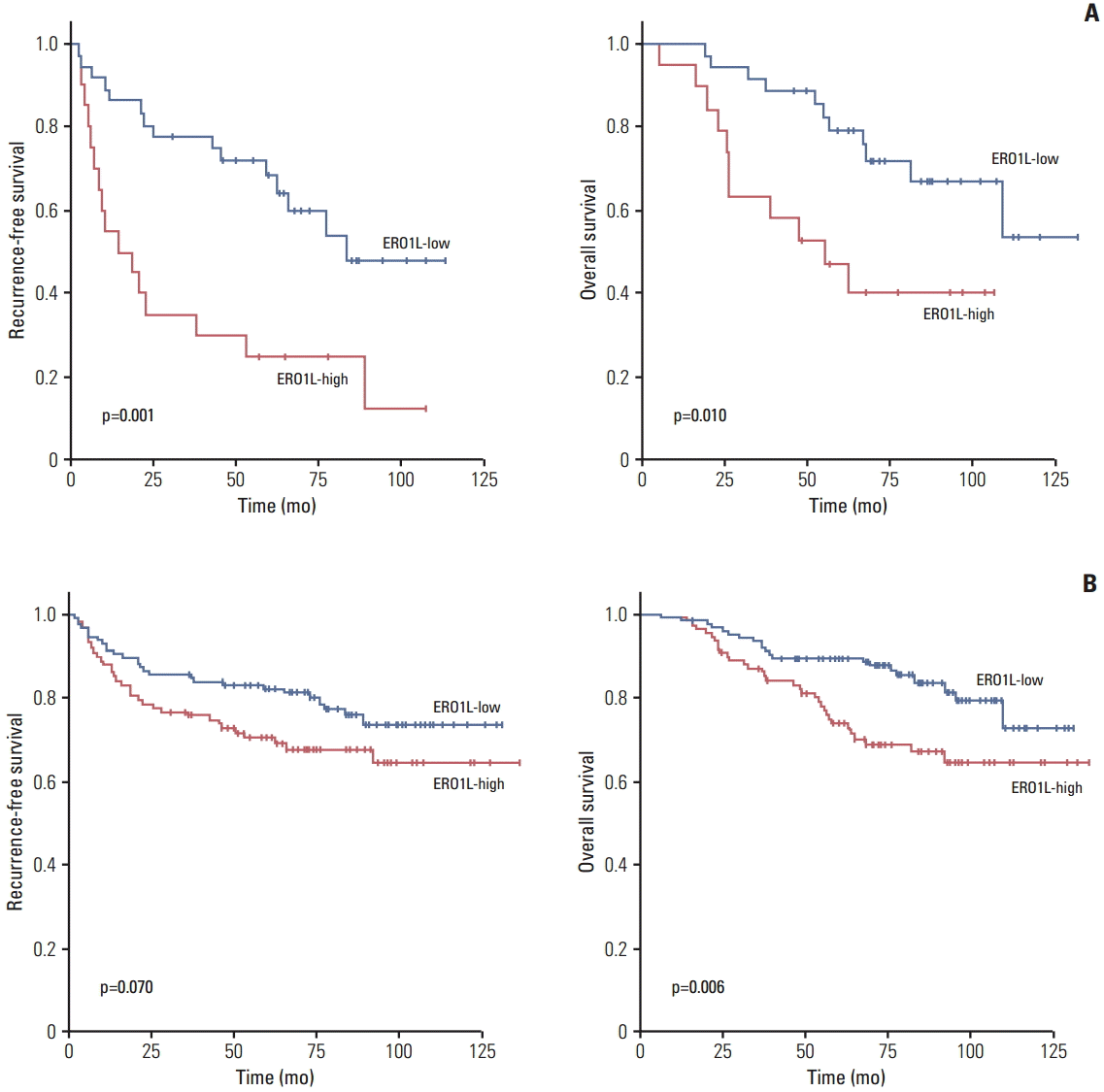 | Fig. 3.Kaplan-Meier plot of recurrence-free survival (RFS) and overall survival (OS) according to gene expression. (A) Gene expression was measured using quantitative reverse transcription polymerase chain reaction in 56 stage III gastric cancer patients. (B) Gene expression was measured using immunohistochemical staining analysis in 231 gastric cancer patients divided into two groups according to their gene expression levels. ERO1L, endoplasmic reticulum oxidoreduction 1-α. |
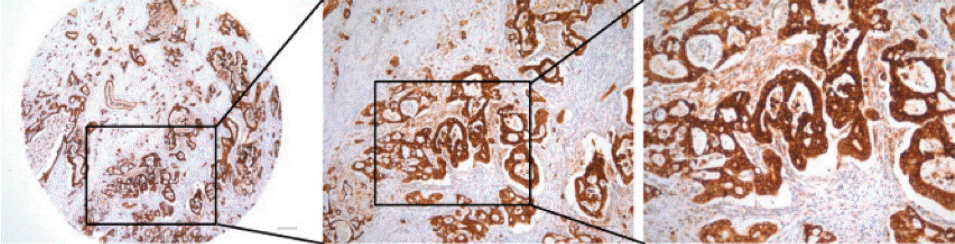 | Fig. 4.Representative images of endoplasmic reticulum oxidoreduction 1-α (ERO1L) protein expression in gastric cancer. High immunohistochemical staining of ERO1L in cytoplasm of gastric cancer (left, ×40; middle, ×100; right, ×200). |
Table 2.
2. ERO1L expressed in human gastric cancer cell lines
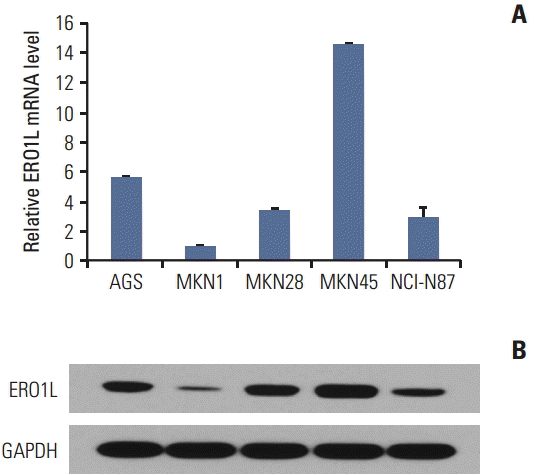 | Fig. 5.Endoplasmic reticulum oxidoreduction 1-α (ERO1L) expression in gastric cancer cell lines. (A) ERO1L mRNA levels were assessed using quantitative real-time reverse transcription polymerase chain reaction. Expression of β-actin was included as an internal loading control. (B) ERO1L protein levels were analyzed using immunoblotting. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was included as an internal loading control. |
3. ERO1L knockdown reduces cell proliferation in gastric cancer cells
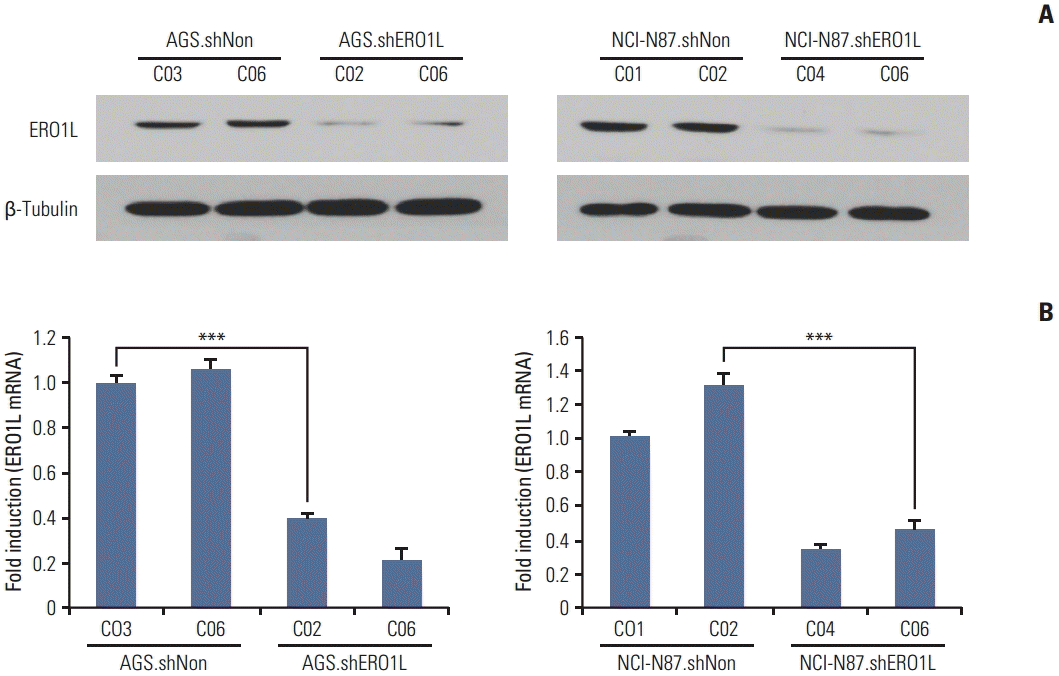 | Fig. 6.Endoplasmic reticulum oxidoreduction 1-α (ERO1L) knockdown expression in lentivirus-mediated stable cells. Total RNA and whole cell lysates were collected from AGS and NCI-N87 cells transfected with shNonTarget and shERO1L vector (AGS, shNon.C03 and C06/shERO1L.C02 and C06; NCI-N87, shNon.C01 and C02/shERO1L.C04 and C06). (A) Expression of ERO1L protein was analyzed by immunoblotting, and mRNA level was measured by quantitative real-time reverse transcription polymerase chain reaction. β-Tubulin and β-actin were included as an internal loading control. ***p < 0.001. |
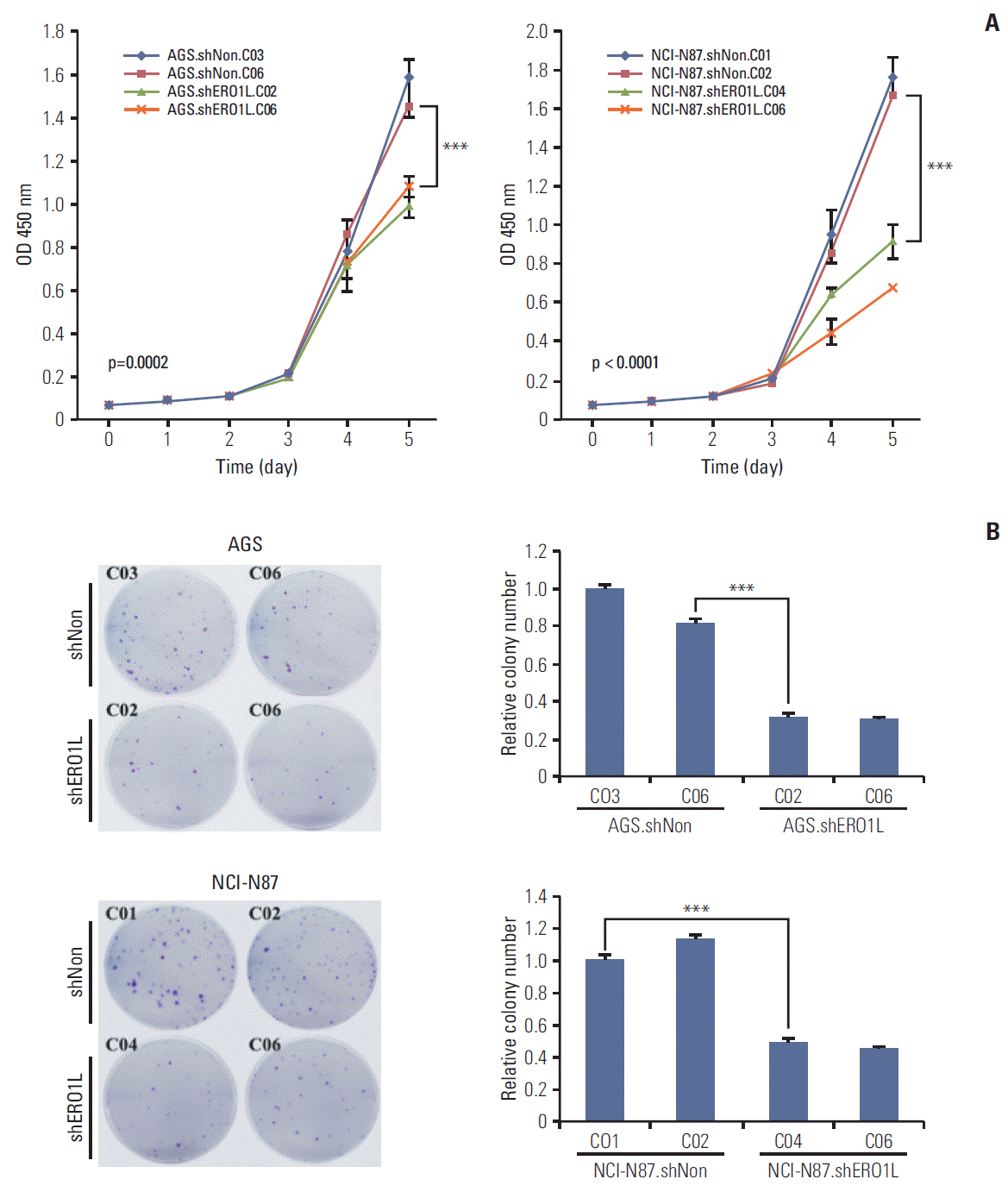 | Fig. 7.Endoplasmic reticulum oxidoreduction 1-α (ERO1L)–deficient gastric cancer cells decrease cell proliferation. (A) Cell proliferation was determined by WST-1 assay. Cell proliferation curves for AGS and NCI-N87 cells at indicated times. Error bars represent mean±standard deviation (SD) of triplicate experiments. (B) A clonogenic assay was performed on AGS and NCI-N87 cells for 21 days. Left panel shows representative images, and right panel shows quantification of colonies. Error bars represent mean±standard deviation of triplicate experiments. ***p < 0.001. |
4. ERO1L expression is associated with gastric cancer cell invasiveness and migration
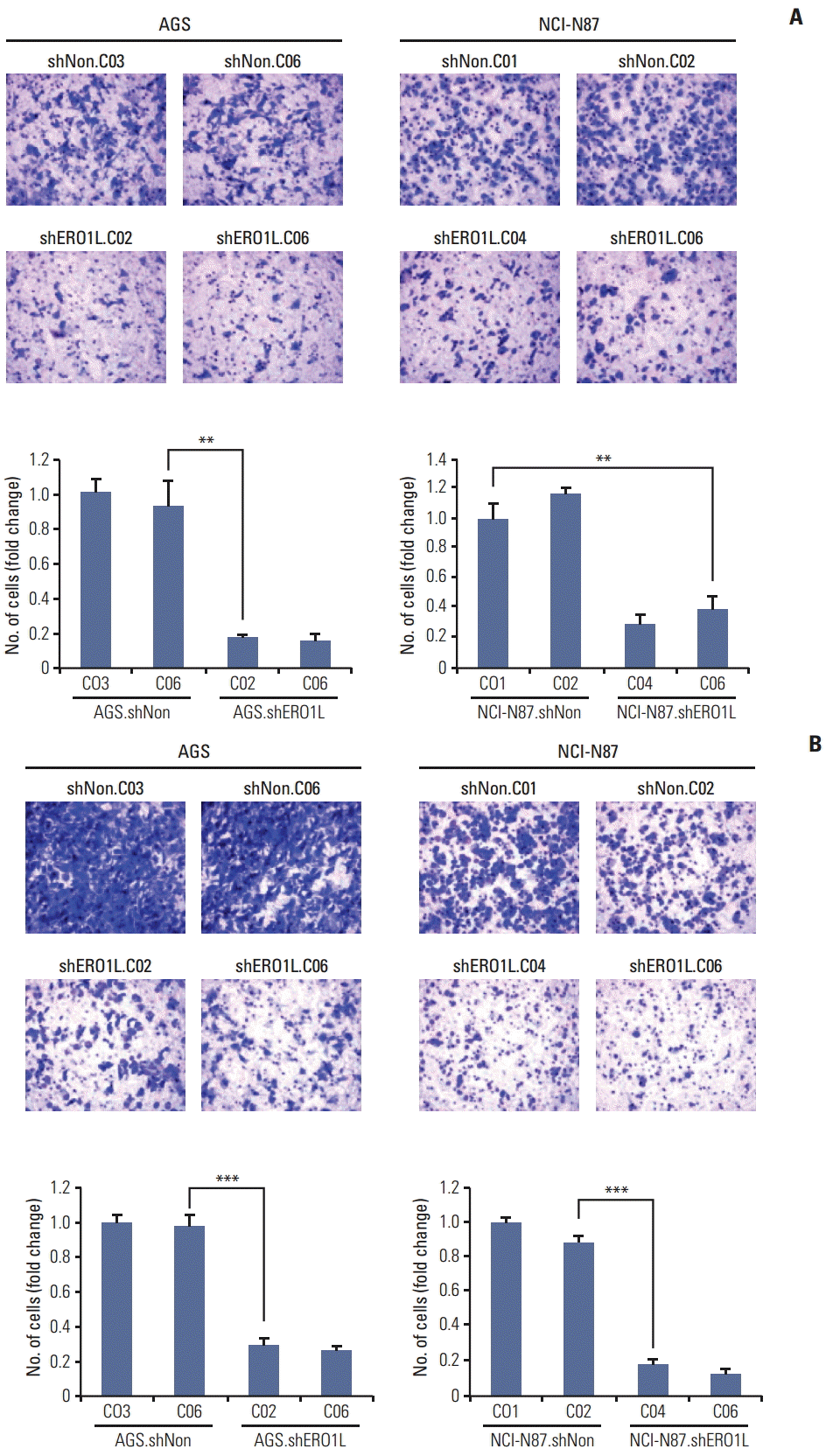 | Fig. 8.Silencing of endoplasmic reticulum oxidoreduction 1-α(ERO1L) expression inhibits migration and invasion abilities of AGS and NCI-N87 cells. (A) Cell migration was determined using Boyden chambers. (B) Cell invasion was assayed using a membrane coated with Matrigel. Five random microscopic fields were counted for each group. The results presented are an average of five random microscopic fields from three independent experiments. **p < 0.01, ***p < 0.001. |
5. ERO1L is related to chemoresistance in gastric cancer
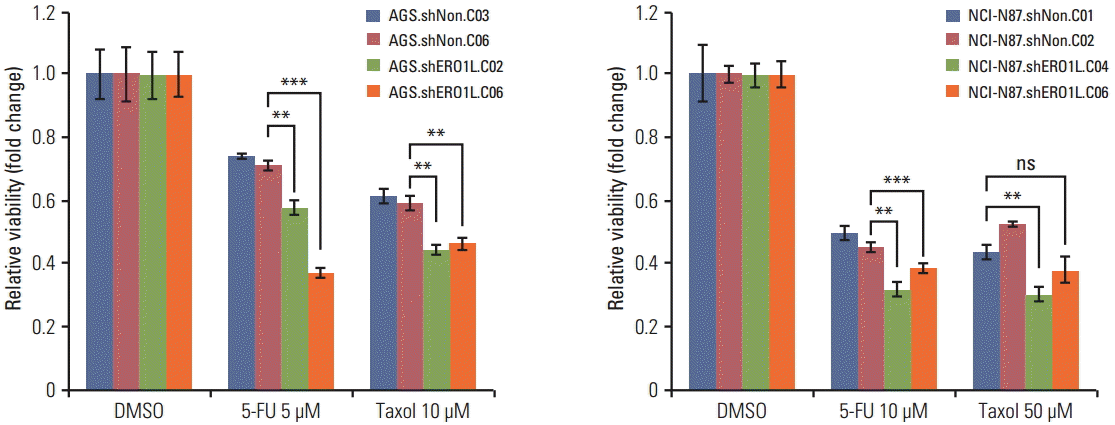 | Fig. 9.Endoplasmic reticulum oxidoreduction 1-α (ERO1L)–deficient gastric cancer cells reduce chemoresistance. Viability of ERO1L-deficient gastric cancer cells treated for 48 hours with chemotherapeutic agents (paclitaxel and 5-fluorouracil [5-FU]) measured by WST-1 assay. Cells treated with dimethylsulfoxide (DMSO) as a control. Data are shown as relative viability (fold change) to the control groups. Data are mean±standard deviation of triplicate experiments. **p < 0.01, ***p < 0.001; ns, not significant. |
6. Down-regulation of AKT and JNK signaling by ERO1L knockdown
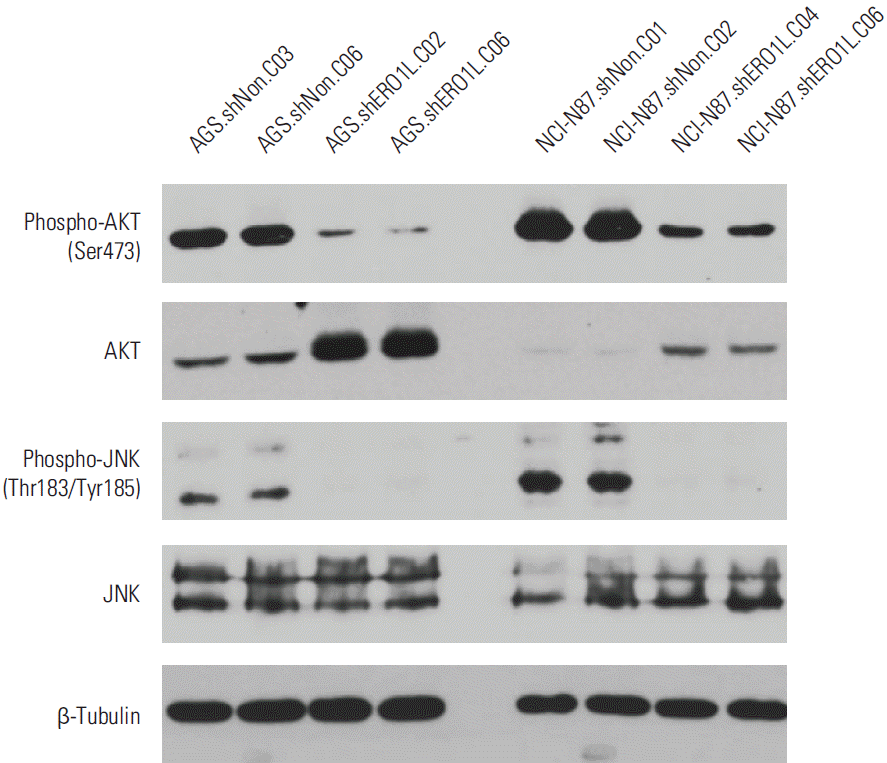 | Fig. 10.Inhibitory effects of endoplasmic reticulum oxidoreduction 1-α (ERO1L)–deficient gastric cancer cells on Akt and JNK signaling pathway. Expression levels of total Akt, phosphorylated Akt at Ser473, total JNK, phosphorylated JNK at Thr183/Tyr185, and β-tubulin (internal control) were measured in the cell lysates. Akt phosphorylation at Ser473 and JNK phosphorylation at Thr183/Tyr185 surprisingly decreased in ERO1L-deficient gastric cancer cells (AGS, shERO1L.C02 and C06; NCI-N87, shERO1L.C04 and C06). |




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download