Abstract
Purpose
The importance of long noncoding RNAs (lncRNAs) in tumorigenesis has recently been demonstrated. However, the role of lncRNAs in development of thyroid cancer remains largely unknown.
Materials and Methods
Using quantitative reverse transcription polymerase chain reaction, expression of three lncRNAs, including BRAF-activated long noncoding RNA (BANCR), papillary thyroid cancer susceptibility candidate 3 (PTCSC3), and noncoding RNA associated with mitogen-activated protein kinase pathway and growth arrest (NAMA), was investigated in the current study.
Results
Of the three lncRNAs (BANCR, PTCSC3, and NAMA), expression of BANCR was significantly up-regulated while PTCSC3 and NAMA were significantly down-regulated in papillary thyroid carcinoma (PTC) compared to that in normal tissue. BANCR-knockdown in a PTC-derived cell line (IHH-4) resulted in significant suppression of thyroid stimulating hormone receptor (TSHR). BANCR-knockdown also led to inhibition of cell growth and cell cycle arrest at G0/G1 phase through down-regulation of cyclin D1. In addition, BANCR was enriched by polycomb enhancer of zeste homolog 2 (EZH2), and silencing BANCR led to decreased chromatin recruitment of EZH2, which resulted significantly reduced expression of TSHR.
Thyroid cancer, the most common malignant tumor of the endocrine organs, has shown steadily increasing morbidity in recent decades [1]. Papillary thyroid carcinoma (PTC) is the main form of nonmedullary thyroid cancer, accounting for approximately 80% of all thyroid cancers [2]. In addition to genetic factors and environmental exposure, recent studies have found that epigenetic alteration may also play an important role in development of variety types of tumors including thyroid cancer. In this regard, involvement of long noncoding RNAs (lncRNAs), particularly BRAF-activated long noncoding RNA (BANCR), papillary thyroid cancer susceptibility candidate 3 (PTCSC3), and noncoding RNA associated with mitogen-activated protein kinase pathway and growth arrest (NAMA), in regulation of thyroid cancer cell survival and apoptosis has been reported [3-5].
LncRNAs utilize a variety of mechanisms in regulation of gene expression including chromatin modification, transcriptional, or post-transcriptional modulation. Through binding to nuclear protein such as polycomb enhancer of zeste homolog 2 (EZH2), a histone methyltransferase, lncRNAs modify histone and by this mechanism, lncRNAs regulate expression of target genes [6,7]. Cis- (stimulate) and trans-regulation (repress) are two main types of transcriptional regulation, under which lncRNAs can target genomically local and distant genes, respectively. Post-transcriptional regulation by lncRNAs includes mRNA splicing, editing, trafficking, translation, and degradation.
Thyroid stimulating hormone (TSH), also known as thyrotropin, is a pituitary hormone that stimulates the thyroid through binding to its receptor (TSHR). TSHR is found mainly on thyroid follicular cells and aberrant expression of TSHR may be involved in development of thyroid cancer as well as Grave’s disease [8-11].
While altered expression of lncRNAs, including BANCR, PTCSC3 and NAMA, has been reported in a variety of cell types of thyroid cancer [3-5], the role of these lncRNAs in modulating TSHR expression, has not been reported. Therefore, the current study was designed to investigate expression of BANCR, PTCSC3, and NAMA in thyroid cancer tissues, and the role of these lncRNAs in regulation of TSHR in a thyroid cancer cell line. Here, we report that three lncRNAs, BANCR, PTCSC3, and NAMA, were differentially expressed in PTC tissue and its adjacent normal tissue. BANCR-knockdown resulted in significantly suppressed proliferation of PTC-derived cell line, IHH-4. In addition, BANCR was enriched by EZH2 and interference of BANCR led to decreased chromatin recruitment of EZH2 and resulted in significant reduction in TSHR expression.
A human PTC-derived cell line, IHH-4, was used for in vitro cell culture experiments. IHH-4 cell line was cultured with RPMI1640 medium supplemented with 10% fetal calf serum at 37°C and 5% CO2 atmosphere. Passages 5 to 10 were used in the current study.
Transfection was performed following the manufacturer’s instruction. Briefly, IHH-4 cells were grown to 75% confluency on 60-mm dishes in RPMI1640 supplemented with 10% fetal calf serum. Lipofectamine-2000 (24 μL; Life Technology, Grand Island, NY) was mixed with 400 μL of serum- and antibiotic-free Opti-MEM (Life Technology) for each dish transfected, and allowed to sit at room temperature for 5 minutes. Simultaneously, 48 μL of each siRNA (GE Dharmacon, Lafayette, CO) was mixed with 800 μL of Opti-MEM (for each dish) and likewise incubated. The two suspensions were then combined and incubated for an additional 20 minutes at room temperature. After washing cell layers gently with phosphate bufferd saline (PBS), 1.2 mL of the mixture with the addition of 0.8 mL Opti-MEM was added to each dish, followed by incubation overnight (18 hours) at 37°C and 5% CO2. Cells were then fed normal growth medium and used for experimentation as designed. Non-targeting control siRNA (GE Dharmacon) was used as a negative control.
Cell proliferation was assessed using a Cell Counting Kit-8 (CCK-8; Beyotime Institute of Biotechnology, Jiangsu, China). Briefly, 24 hours after transfection with control-siRNA or BANCR-siRNA, cells were trypsinized and plated in triplicate wells of a 96-well plate at a density of 2×103 cells/well. Cells were allowed to grow in RPMI1640 supplemented with 10% fetal calf serum for 5 days. The number of cells per well was determined by the absorbance at 450 nm at indicated time points, following the manufacturer’s instructions. Data were expressed as value of optical density at 450 nm wave length.
Clonogenic assay was performed with a modification of a previously published method [12]. Briefly, after completion of siRNA transfection, the cells were plated into 60-mm tissue culture dishes in triplicate and at a cell density of 100 cells/dish. Cells were then allowed to grow in RPMI1640 supplemented with 10% fetal calf serum for 14 days. Cells were then fixed and stained with Diff-Quick. After air drying, colonies were counted under a microscope and cell number in each colony was at least 20 cells.
After transfection with siRNA, cells were cultured in RPMI1640 supplemented with 10% fetal calf serum for an additional 48 days. Cells were then trypsinized and suspended with RPMI1640 containing 10% serum. Cells were then pelleted, suspended with cold ethanol/PBS at a ratio of 2.5:1 and fixed at 4°C for at least 30 minutes. Cells were then stained with propidium iodide solution for 30 minutes as described previously [13]. Cell cycle and DNA content were then analyzed by flow cytometry.
Total RNA from cells or tissue was extracted using Trizol (Life Technology). After RNA quantification with Nanodrop 1000 (Thermo Scientific, Wilmington, DE), reverse transcription was performed using a commercially available kit (Applied Biosystem, Life Technology, Grand Island, NY) following the manufacturer’s instructions. Quantitative real time polymerase chain reaction (PCR) was then performed using commercially available pre-synthesized probe/primers and reagents, and the 7500 Real Time System (Applied Biosystems, Life Technology) following the manufacturer’s instructions. Each reaction was performed in triplicate, and the 2-ΔΔCt method was used to determine the relative gene expression levels. As an internal control, 18s RNA was used to normalize the data.
Proteins of total cell lysate or proteins from the tissue were extracted using RIPA lysis and extraction buffer. After brief sonication and centrifugation, protein concentration was determined using a Bio-Rad protein assay kit (Bio-Rad, Hercules, CA). A total of 10 μg protein was electrophoresed on a 10% sodium dedecyl sulfate polyacryamide gel and subsequently transferred to a polyvinylidene difluoride membrane (Bio-Rad). The membrane was blocked with 5% skimmed milk in Tris-buffered saline containing 0.1% Tween-20 (TBST) at room temperature for 1 hour. The membrane was then incubated with the desired primary antibodies (1:1,000 dilution) at 4°C overnight. The next day, the membrane was washed 3 times with TBST, followed by incubation with horseradish peroxidase-conjugated secondary anti-rabbit or anti-mouse antibody (1:4,000, Rockland, Limerick, PA) at room temperature for 1 hour. After washing 3 times with TBST, the membrane was developed using ECL Plus (EMD Millipore, Billerica, MA), and exposed to X-ray film. β-Actin was used as an internal loading control.
Chromatin immunoprecipitation (ChIP) assay was performed using an EZ ChIP Chromatin Immunoprecipitation Kit (Millipore, Bedford, MA) following the manufacturer’s instructions. Briefly, cross-linked chromatin was sonicated into 200-bp to 1,000-bp fragments. The chromatin was immunoprecipitated using anti-EZH2 antibody (Cell Signaling Technology, Beverly, MA). Normal mouse IgG was used as a negative control. Quantitative PCR was then performed using SYBR Green Mix (Takara Bio, Otsu, Japan).
RNA immunoprecipitation (RIP) assay was performed using a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) following the manufacturer’s instructions. The anti-EZH2 antibody and IgG (as control) was used for RIP (Cell Signaling Technology). Co-precipitated RNAs were detected by quantitative reverse transcription polymerase chain reaction (RT-PCR). Total RNAs (input controls) and isotype controls were assayed simultaneously to demonstrate that the detected signals were the result of RNAs specifically binding to EZH2.
With informed consent, a total of 40 pairs of specimens of human PTC and adjacent normal tissue were obtained from surgical procedures performed at Yantai Yuhuangding Hospital from September 2013 to November 2014 (Yantai, Shandong, China). The protocol of the current study was approved by the Protection of Human Ethics Committee, College of Medicine, Qingdao University (Qingdao, Shandong, China). PTC diagnosis was pathologically confirmed, tissues were snap frozen with liquid nitrogen, and stored at –80°C till RNA extraction.
Data were analyzed using PRISM 6 software (GraphPad, La Jolla, CA), and expressed as mean±standard deviation of the data from at least three separate experiments. Paired data were analyzed by Student’s t test, and group data were analyzed by one-way analysis of variance (ANOVA) followed by Tukey correction, as appropriate. p < 0.05 was considered significantly different in comparison.
A total of 40 pairs of PTC tissue and its adjacent normal tissue were obtained from clinically ongoing surgical specimens with informed consent. Cellular types of the PTCs were PTC with poorly differentiated area (6 cases), follicular variant (15 cases), diffuse sclerosing variant (8 cases), and CP classical PTC (21 cases). To examine the role of lncRNAs in the pathogenesis of thyroid carcinoma, expression of three lncRNAs, BANCR, PTCSC3, and NAMA in these tumor tissues as well as their adjacent normal tissues was assessed by quantitative real-time RT-PCR. Interestingly, BANCR expression was significantly up-regulated in PTC compared to that in its adjacent normal tissue (p < 0.0001) (Fig. 1A). In contrast, expression of PTCSC3 and NAMA was significantly down-regulated in PTC compared to that in normal tissue (p < 0.0001 and p=0.0002, respectively) (Fig. 1B and C).
Since thyroid cell proliferation and function is mediated by the interaction of TSH and its receptor (TSHR), the effect of silencing lncRNA on TSHR expression was investigated using a cell line of PTC, IHH-4 cells. For suppression of BANCR, PTCSC3, and NAMA, siRNAs specifically targeting BANCR, PTCSC3, or NAMA were transiently transfected. Efficiency of lncRNA silence was determined by real-time RT-PCR and at least 60% suppression was demonstrated (data not shown). Silencing BANCR resulted in significant down-regulation of TSHR (p < 0.01) (Fig. 2A), while neither PTCSC3-siRNA, nor NAMA-siRNA had a significant effect on TSHR expression in IHH-4 cells (Fig. 2B and C, respectively).
Following transfection of BANCR-siRNA, IHH-4 cells were allowed to grow in full medium for 5 days. Growth of cells transfected with BANCR-siRNA was significantly slower than that of cells with control-siRNA on day 3 and day 5 (p < 0.05 and p < 0.01, respectively) (Fig. 3A). A clonogenic assay was performed to confirm the effect of BANCR suppression on cell proliferation. Consistent with the result of cell proliferation assay, number of clones was significantly decreased in cells lacking BANCR (200.3±25.5 of controlsiRNA vs. 56.7±8.5 of BANCR-siRNA, p < 0.01) (Fig. 3B).
To further investigate the role of BANCR in regulating cell proliferation, cellular DNA content and cell cycle profiling was performed. As shown in Fig. 4A and B, G1 to S phase transition was significantly decreased. Gene expression and protein level of cyclin proteins including cyclin D1, cyclin E1, cyclin A1, and cyclin B1 were also determined following BANCR-siRNA transfection into IHH-4 cells. Of particular interest, cyclin D1 mRNA and protein was significantly decreased in cells transfected with BANCR-siRNA compared to that of control-siRNA transfected cells (p < 0.05) (Fig. 4C and D). In contrast, mRNAs of cyclin A1, E1, or B1 were not altered in cells after silencing BANCR (Fig. 4C), although proteins of cyclin A1 and E1 were slightly but not significantly reduced in cells lacking BANCR (Fig. 4D).
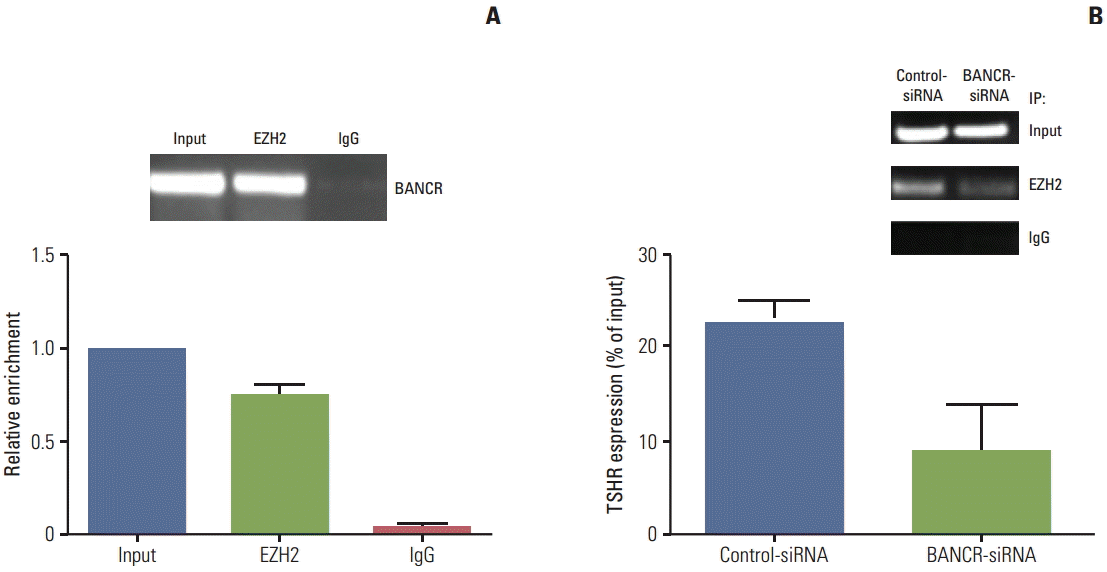
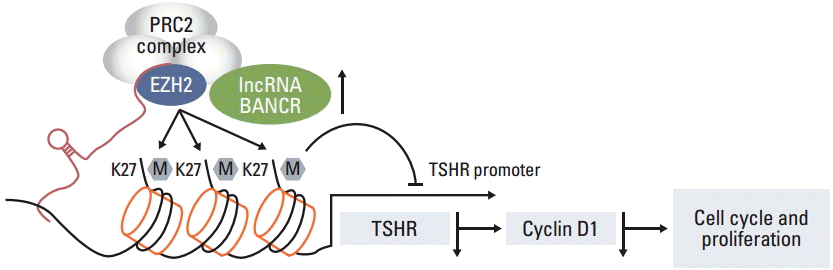
EZH2 is known to inhibit genes responsible for suppressing tumor growth, and lncRNAs modulate tumor cell growth through binding to EZH2 in variety of cancer cells [14,15], therefore, association of BANCR and EZH2 was further investigated in the current study. First, RIP assay was performed using anti-EZH2 and IgG as a negative control. Then, quantitative RT-PCR was performed for quantification of the retrieved RNA. As shown in Fig. 5A, BANCR was highly enriched in the EZH2 group compared to the non-immune IgG group. Next, ChIP assay was performed to further determine whether BANCR knockdown altered expression of TSHR. As shown in Fig. 5B, expression of TSHR was significantly decreased in BANCR-knockdown cells compared to control cells, suggesting that BANCR may enhance TSHR expression through binding to EZH2.
Accumulating evidence suggests that lncRNAs may play an important role in development of various types of cancer. PTC is the most common type of thyroid cancer. In the current study, we first examined expression of three lncRNAs in the PTC and its adjacent normal tissue. It was found that BANCR was significantly up-regulated in PTC tissue compared to normal tissue, and in contrast, expression of PTCSC3 and NAMA was significantly decreased in tumor tissue. Using in vitro cell culture and molecular strategy, we further demonstrated that BANCR regulated expression of TSHR, while neither PTCSC3 nor NAMA had an effect on TSHR expression. BANCR controlled growth and cell cycle of IHH-4 cells (a cell line of PTC) through regulation of cyclin D1 protein expression. In addition, physical binding of BANCR to EZH2 was demonstrated by RIP assay, and by this mechanism, BANCR may regulate TSHR as well as subsequent target genes including cyclin D1 (Fig. 6).
PTC is the main form of nonmedullary thyroid cancer, accounting for approximately 80% of all thyroid cancers. In contrast to other cancers, the incidence of PTC has increased in recent decades, but this may due to improved diagnostics [2]. Although the etiology of PTC is not well defined, it is clearly influenced by genetic factors and environmental exposures. Radiation exposure of the thyroid during childhood is the most clearly defined environmental factor. However, genetic predisposition also plays an important role as evidenced by case control studies [16]. In addition, recent accumulating evidence indicates that epigenetic alteration may also play an important role in the development of various kinds of tumors including thyroid cancers. In this context, involvement of lncRNAs, particularly BANCR, PTCSC, and NAMA, in regulation of thyroid cancer cell survival and apoptosis has been reported [3-5].
Noncoding RNAs are a group of RNAs that do not code for protein synthesis. They are categorized according to two classes based on transcript size: “long noncoding RNAs (lncRNAs)” are greater than 200 nucleotides in length, and “small noncoding RNAs” are less than 200 nucleotides in length including microRNAs [17,18]. Unlike miRNAs, which have been studied extensively in the last decade, lncRNAs are more diverse, not highly conserved, and have been regarded as transcriptional “noise” and not biologically functional for a long period of time [19,20]. Recently, however, accumulating evidence has revealed the significance of lncRNAs in regulation of multiple biological functions at all levels of chromosome modification, mRNA transcription, and post-transcriptional protein synthesis [21-23]. Notably, several lncRNAs including BANCR, PTCSC3, and NAMA have been reported to play an important role in regulating thyroid cancer formation and progression [3-5]. Consistent with previous reports, we found that expression of BANCR was significantly up-regulated in PTC compared to that in normal tissue, while expression of PTCSC3 and NAMA was dramatically down-regulated in PTC compared to its adjacent normal tissue, suggesting that thyroid carcinogenesis is positively regulated by BANCR and negatively regulated by PTCSC3 and NAMA.
In the current study, biological function of BANCR in thyroid cancer was further investigated utilizing an in vitro IHH-4 cell culture. IHH-4, a cell line derived from PTC, has been used by other investigators in study of thyroid cancer biology [3]. Consistent with findings in human PTC tissue, IHH-4 cells also highly express BANCR (data not shown). In this study, silencing BANCR with siRNA in IHH-4 cells resulted in remarkable suppression of cell proliferation, and cell cycle arrest at G0/G1 phase. In addition, significant down-regulation of both mRNA and protein of cyclin D1 was observed in IHH-4 cells lacking BANCR. Paradoxically, mRNA of cyclin A1, E1, and B1 was not altered in cells transfected with BANCR-siRNA, although proteins of cyclin A1 and E1 were slightly but not significantly reduced in cells lacking BANCR, suggesting that BANCR regulated cell growth and cell cycle through modulation of cyclin D1.
TSH exerts its effect on thyroid cell proliferation through binding to its receptor, TSHR. TSHR is mainly found on thyroid follicular cells. According to some reports, pathogenesis of thyroid cancer and Grave’s disease may be attributed to the aberrant expression of TSHR [8-11]. While we were unable to examine expression of TSHR in PTC tissues due to lack of sources, which is a pitfall of the current study, we demonstrated that BANCR-knockdown in IHH-4 cells resulted in a significant reduction in the TSHR protein. Interestingly, however, TSHR protein synthesis was not significantly altered by either PTCSC3-siRNA or NAMA-siRNA, although TSHR protein was slightly decreased in cells transfected with NAMA-siRNA, indicating that BANCR may contribute to development of PTC through regulation of TSHR, but not PTCSC3 or NAMA.
Tumorigenesis and progression of cancer result not only from genetic factors and environmental exposure, but also from epigenetic alteration. Epigenetic modifications include DNA methylation, histone modifications, and modulation by miRNAs or lncRNAs. Histone modification has been reported to play a role in the tumorigenesis of thyroid cancer, and EZH2, a histone methyltransferase, is a well-known histone modifier. In this regard, researchers have demonstrated a strong association of overexpression of EZH2 with progression and invasion of several types of cancers including thyroid cancer [7,14,15]. In addition, studies have shown that EZH2 was up-regulated by aberrant expression of MYC transcription factor and miRNAs such as miR-26a [24,25]. Interaction of BANCR and EZH2 in the tumorigenesis of thyroid cancer, however, has not been reported. The results of the current study demonstrated that BANCR was enriched by EZH2, and that silencing BANCR by siRNA resulted in decreased chromatin recruitment of EZH2 as well as significant reduction in TSHR expression as evidenced by ChIP assay with anti-EZH2 antibody, suggesting that BANCR could stimulate TSHR expression by transcriptional up-regulation of the TSHR promoter through EZH2 enrichment. The current study, however, did not determine whether histone methylation in the promoter region of cyclin D1 was affected by the suppression of BANCR and reduced chromatin recruitment of EZH2, which is a limitation of the current study and remains to be defined in future study.
Taken together, the three lncRNAs tested in the current study were differentially expressed in PTC tissue and a PTC-derived cell line (IHH-4). Specifically, BANCR was significantly up-regulated in PTC, and silencing BANCR with siRNA resulted in dramatically reduced expression of TSHR as well as suppression of cell proliferation via a mechanism of cell cycle arrest. In addition, BANCR was enriched by EZH2 and BANCR was required for EZH2 binding to TSHR promoter because interference of BANCR abrogated chromatin recruitment of EZH2 and resulted in significant reduction of TSHR expression. These findings suggested regulation of BANCR as a therapeutic goal of PTC.
The current study demonstrated that BANCR was significantly up-regulated in PTC, and silencing BANCR with siRNA resulted in dramatically reduced expression of TSHR as well as suppression of cell proliferation via a mechanism of cell cycle arrest. These findings indicated that regulation of BANCR be a therapeutic strategy for PTC.
References
1. Davies L, Welch HG. Increasing incidence of thyroid cancer in the United States, 1973-2002. JAMA. 2006; 295:2164–7.

2. Davies L, Welch HG. Current thyroid cancer trends in the United States. JAMA Otolaryngol Head Neck Surg. 2014; 140:317–22.

3. Wang Y, Guo Q, Zhao Y, Chen J, Wang S, Hu J, et al. BRAF-activated long non-coding RNA contributes to cell proliferation and activates autophagy in papillary thyroid carcinoma. Oncol Lett. 2014; 8:1947–52.

4. Fan M, Li X, Jiang W, Huang Y, Li J, Wang Z. A long non-coding RNA, PTCSC3, as a tumor suppressor and a target of miRNAs in thyroid cancer cells. Exp Ther Med. 2013; 5:1143–6.

5. Yoon H, He H, Nagy R, Davuluri R, Suster S, Schoenberg D, et al. Identification of a novel noncoding RNA gene, NAMA, that is downregulated in papillary thyroid carcinoma with BRAF mutation and associated with growth arrest. Int J Cancer. 2007; 121:767–75.
6. Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004; 18:2627–38.

7. Borbone E, Troncone G, Ferraro A, Jasencakova Z, Stojic L, Esposito F, et al. Enhancer of zeste homolog 2 overexpression has a role in the development of anaplastic thyroid carcinomas. J Clin Endocrinol Metab. 2011; 96:1029–38.

8. He H, Li W, Liyanarachch S, Jendrzejewski J, Srinivas M, Davuluri RV, et al. Genetic predisposition to papillary thyroid carcinoma: involvement of FOXE1, TSHR, and a novel lincRNA gene, PTCSC2. J Clin Endocrinol Metab. 2015; 100:E164–72.

9. Bufalo NE, Dos Santos RB, Marcello MA, Piai RP, Secolin R, Romaldini JH, et al. TSHR intronic polymorphisms (rs179247 and rs12885526) and their role in the susceptibility of the Brazilian population to Graves' disease and Graves' ophthalmopathy. J Endocrinol Invest. 2015; 38:555–61.

10. Khan MS, Pandith AA, Masoodi SR, Wani KA, Ul Hussain M, Mudassar S. Epigenetic silencing of TSHR gene in thyroid cancer patients in relation to their BRAF V600E mutation status. Endocrine. 2014; 47:449–55.

11. Huth S, Jaeschke H, Schaarschmidt J, Paschke R. Controversial constitutive TSHR activity: patients, physiology, and in vitro characterization. Horm Metab Res. 2014; 46:453–61.

12. Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006; 1:2315–9.

13. Telford WG, King LE, Fraker PJ. Rapid quantitation of apoptosis in pure and heterogeneous cell populations using flow cytometry. J Immunol Methods. 1994; 172:1–16.

14. Ngollo M, Lebert A, Dagdemir A, Judes G, Karsli-Ceppioglu S, Daures M, et al. The association between histone 3 lysine 27 trimethylation (H3K27me3) and prostate cancer: relationship with clinicopathological parameters. BMC Cancer. 2014; 14:994.

15. Esposito F, Tornincasa M, Pallante P, Federico A, Borbone E, Pierantoni GM, et al. Down-regulation of the miR-25 and miR-30d contributes to the development of anaplastic thyroid carcinoma targeting the polycomb protein EZH2. J Clin Endocrinol Metab. 2012; 97:E710–8.

16. Dong C, Hemminki K. Modification of cancer risks in offspring by sibling and parental cancers from 2,112,616 nuclear families. Int J Cancer. 2001; 92:144–50.

18. Brosnan CA, Voinnet O. The long and the short of noncoding RNAs. Curr Opin Cell Biol. 2009; 21:416–25.

19. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009; 136:629–41.

20. Jia H, Osak M, Bogu GK, Stanton LW, Johnson R, Lipovich L. Genome-wide computational identification and manual annotation of human long noncoding RNA genes. RNA. 2010; 16:1478–87.

21. Flynn RA, Chang HY. Active chromatin and noncoding RNAs: an intimate relationship. Curr Opin Genet Dev. 2012; 22:172–8.

22. Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013; 193:651–69.

23. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009; 10:155–9.

24. Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008; 322:1695–9.

25. Koh CM, Iwata T, Zheng Q, Bethel C, Yegnasubramanian S, De Marzo AM. Myc enforces overexpression of EZH2 in early prostatic neoplasia via transcriptional and post-transcriptional mechanisms. Oncotarget. 2011; 2:669–83.
Fig. 1.
Expression of long noncoding RNAs (lncRNAs) in papillary thyroid carcinoma (PTC) in comparison to adjacent normal tissue. Total RNA was extracted from PTC tissue and its adjacent normal tissue, and expression of BRAF-activated long noncoding RNA (BANCR) (A), papillary thyroid cancer susceptibility candidate 3 (PTCSC3) (B), and noncoding RNA associated with mitogen-activated protein kinase pathway and growth arrest (NAMA) (C) was quantified as described in the methods. 18s RNA was used as an internal control and data were expressed as relative expression of the lncRNA in the tumor versus respective adjacent normal tissue. n=40 each comparison. Student’s t test was used for comparison of statistical difference.
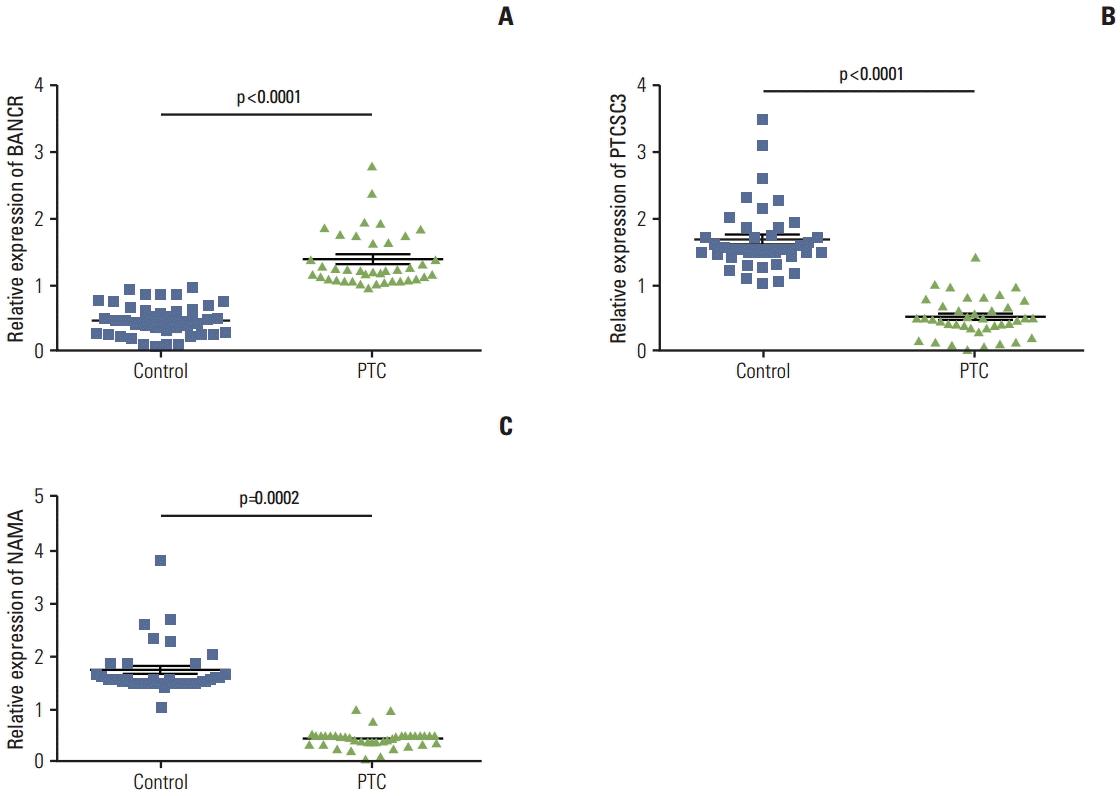
Fig. 2.
Effect of lncRNAs on TSHR expression in in vitro cell culture. A PTC cell line (IHH-4) was transfected with either control-siRNA or a siRNA targeting BANCR (BANCR-siRNA) (A), PTCSC3 (PTCSC3-siRNA) (B), or NAMA (NAMA-siRNA) (C) as described in the methods. Total cell lysate was harvested after 48 hours additional culture. Expression of TSHR protein level was assessed by immunoblotting (inserts) followed by density analysis. β-Actin was used as a loading control. Data presented was an average of three separate experiments. lncRNAs, long noncoding RNAs; TSHR, thyroid stimulating hormone receptor; PTC, papillary thyroid carcinoma; BANCR, BRAF-activated long noncoding RNA; PTCSC3, papillary thyroid cancer susceptibility candidate 3; NAMA, noncoding RNA associated with MAP kinase pathway and growth arrest.
*p < 0.01 compared to control-siRNA treated cells.
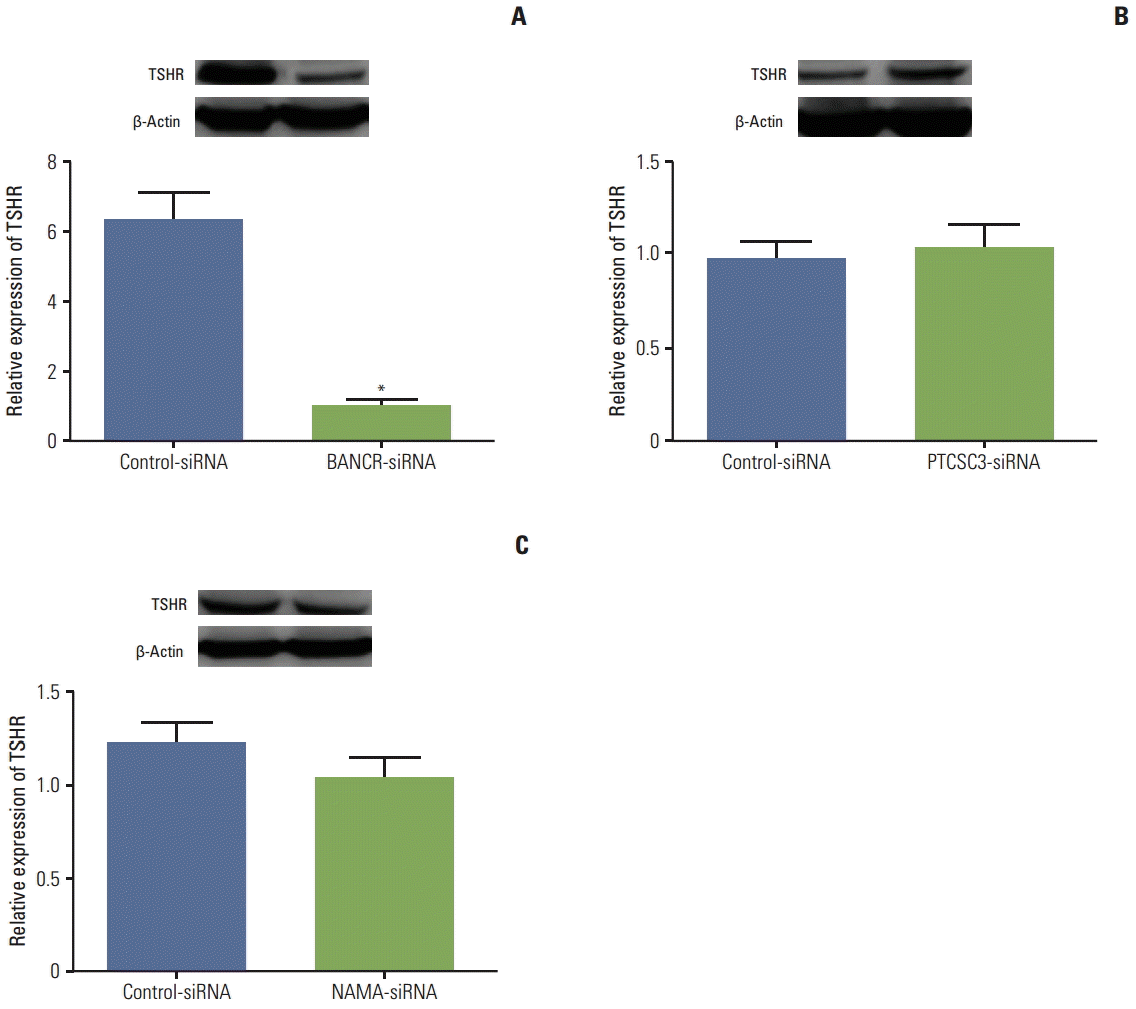
Fig. 3.
Effect of BANCR suppression on cell proliferation. A PTC cell line (IHH-4) was transfected with either control-siRNA or a siRNA targeting BANCR (BANCR-siRNA), cell proliferation was then assessed. (A) Cell proliferation assay by CCK-8. Following siRNA transfection, cells were plated into 96-well plates and allowed to grow for 5 days. Cell number was counted using a commercial kit (CCK-8) as described in the methods. *p < 0.05, **p < 0.01 by multi-group t test. (B) Clonogenic assay. Following transfection with siRNAs, cells were plated into 60-mm dishes and allowed to grow for 14 days and the number of colonies was counted as described in the methods. Data presented was representative of three separate experiments. BANCR, BRAF-activated long noncoding RNA; PTC, papillary thyroid carcinoma.
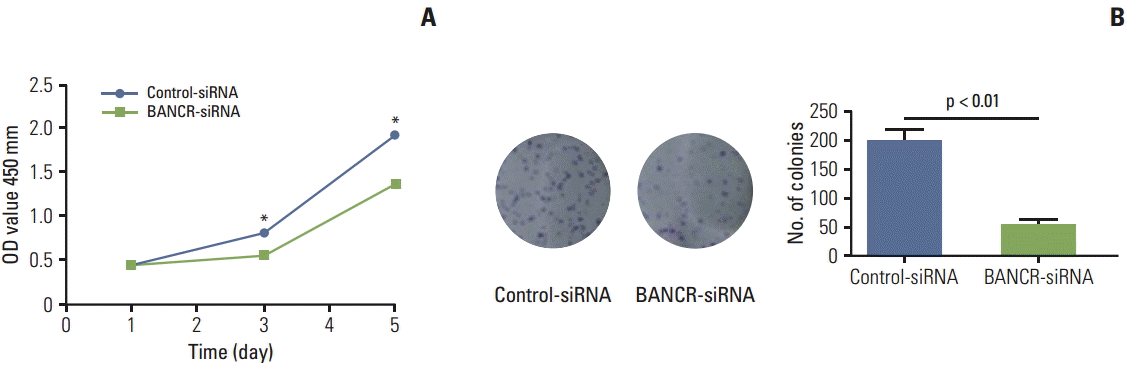
Fig. 4.
Effect of BANCR suppression on expression of cyclin proteins. (A, B) Cell cycle analysis by flow cytometry. IHH-4 cells were transfected with control-siRNA or a siRNA targeting BANCR (BANCR-siRNA). Following transfection with siRNAs, cells were allowed to grow for 48 hours and then trypsinized. After fixation and staining with propidium iodide, cell cycle and DNA content were analyzed by flow cytometry. Data presented was representative (analysis of triplicate samples) of three separate experiments. (C) mRNA expression. Total RNA was extracted 24 hours after completing transfection and mRNA expression of indicated cyclins was quantified by real time RT-PCR. (D) Immunoblotting for cyclin proteins. Cell lysate was harvested 48 hours after completing transfection, and levels of cyclin proteins were assessed by immunoblot as described in the methods. β-Actin was used as a loading control. Data presented was representative of three separate experiments. BANCR, BRAF-activated long noncoding RNA; RT-PCR, reverse transcription polymerase chain reaction.
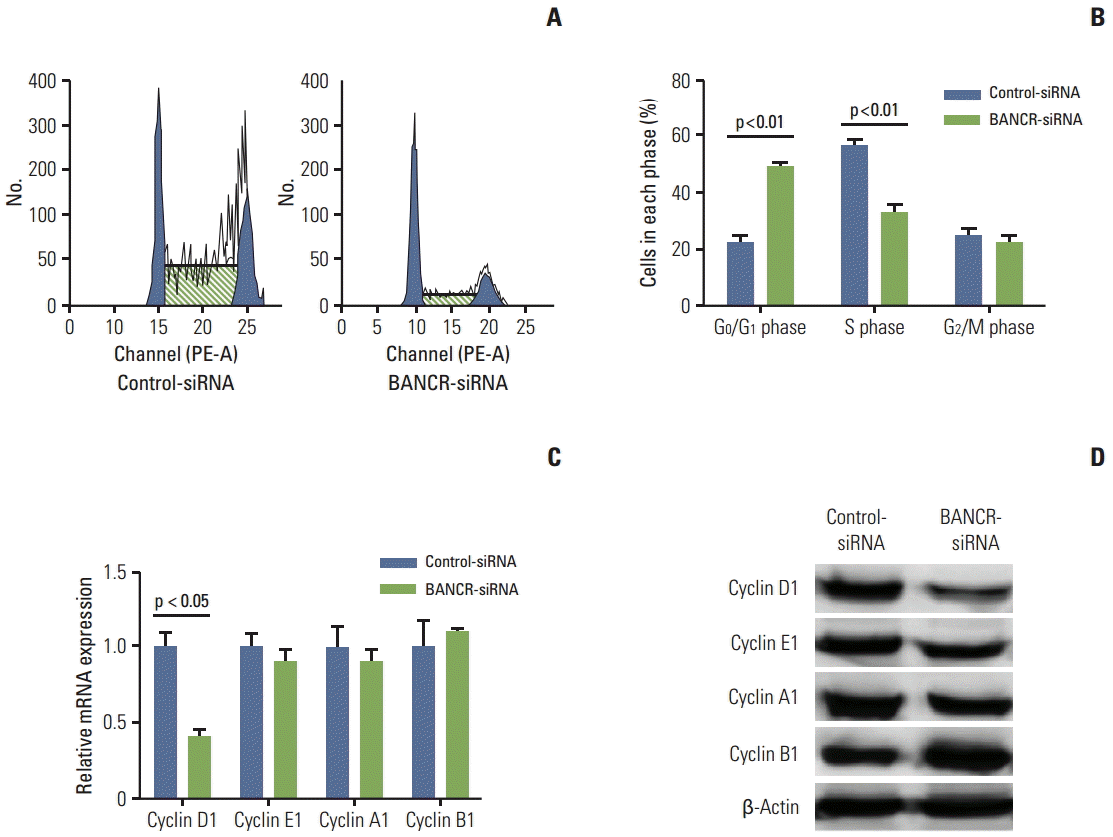
Fig. 5.
Role of EZH2 in mediating BANCR regulation on TSHR. (A) RIP assay. Quantitative RT-PCR analysis of lncBANCR in the RIP from IHH-4 cells was performed using anti-EZH2 antibody and IgG as a negative control. Data presented was representative of three separate experiments. (B) ChIP assay. ChIP assay for IHH-4 cells transfected with control-siRNA or BANCR-siRNA was performed using anti-EZH2 antibody. Semi-quantitative polymerase chain reaction analysis was performed for amplification of associated DNA using primers specific for the TSHR promoter region. IgG indicated the negative control of immunoprecipitation. EZH2, enhancer of zeste homolog 2; BANCR, BRAF-activated long noncoding RNA; TSHR, thyroid stimulating hormone receptor; RIP, RNA immunoprecipitation; RT-PCR, reverse transcription polymerase chain reaction; ChIP, chromatin immunoprecipitation.




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download