Introduction
Materials and Methods
1. Cell culture
2. Plasmids and transfection
3. Antibodies
4. Cell synchronization
5. shRNA plasmid construction
6. Immunoprecipitation and mass spectrometry
7. Immunofluorescence
Results
1. Establishment of the interactome of HP1γ
Fig. 1.
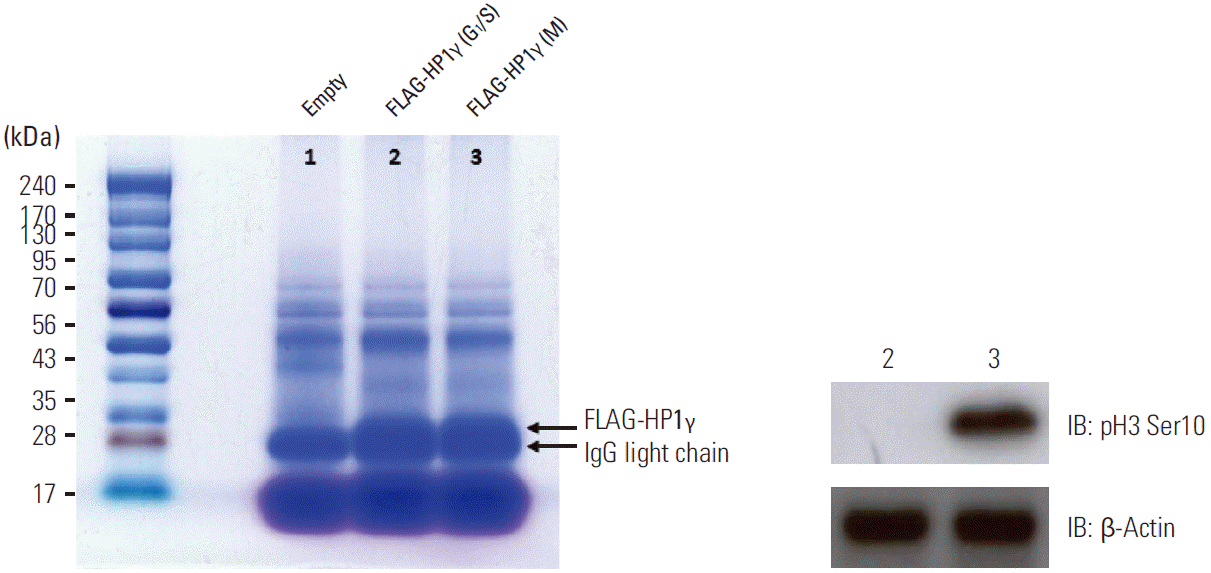
Fig. 2.
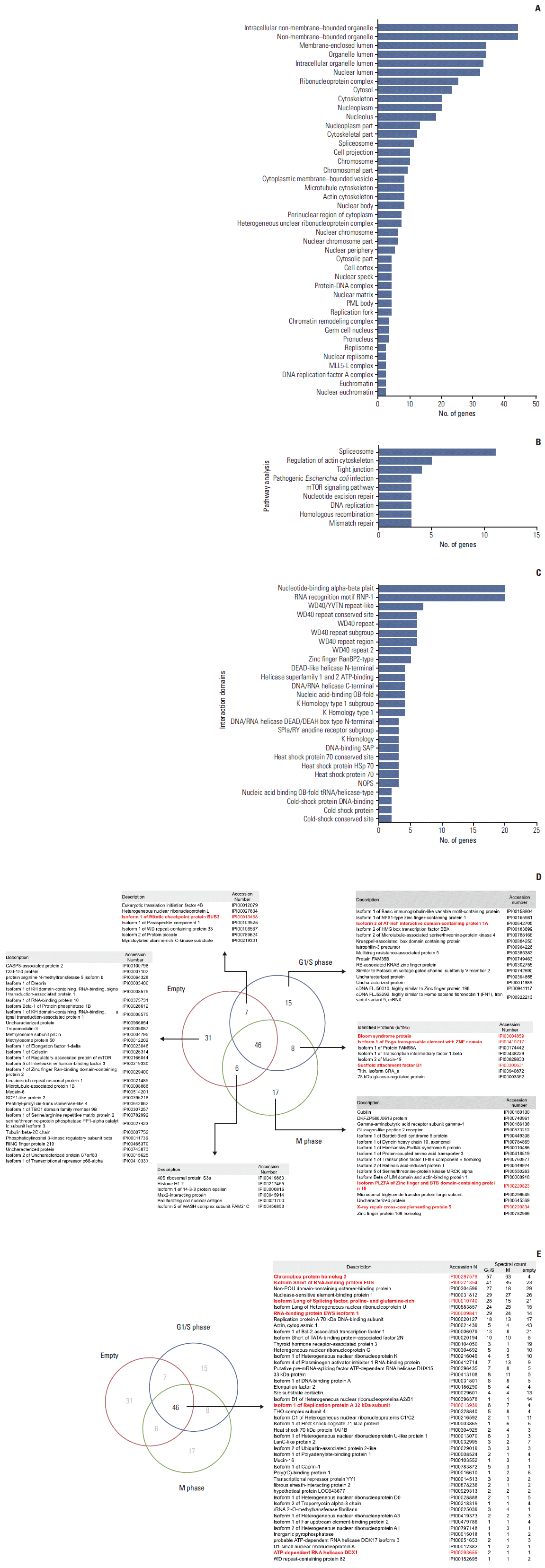
2. The function of HP1γ in the DNA damage response pathway
Table 1.
| Description | Accession No. | M. W. (kDa) |
Unique spectral count |
Significance validation |
|||||
|---|---|---|---|---|---|---|---|---|---|
| G1-S | M | Empty | Fold ratio (G1-S or M/control) | GO | Article references | Co-IP Ex. | |||
| Bloom syndrome protein (BLM) | IPI00004859 | 159 | 2 | 1 | 0 | O | O | - | O |
| Chromobox protein homolog 3 (CBX3, the bait in this study) | IPI00297579 | 21 | 57 | 63 | 4 | O | O | - | Not done |
| Isoform 1 of Replication protein A 32 kDa subunit (RPA2) | IPI00013939 (+2) | 29 | 6 | 7 | 4 | O | O | - | Not done |
| Isoform 2 of AT-rich interactive domain-containing protein 1A (ARID1A) | IPI00642705 (+2) | 218 | 2 | N.D. | N.D. | O | O | - | Not done |
| Isoform Long of splicing factor, proline- and glutamine-rich (SFPQ) | IPI00010740 | 76 | 28 | 15 | 21 | O | O | - | Not done |
| Scaffold attachment factor B1 (SAFB) | IPI00300631 (+5) | 103 | 5 | 1 | N.D. | O | O | - | Not done |
| X-ray repair cross-complementing protein 5 (XRCC5) | IPI00220834 | 83 | N.D. | 2 | N.D. | O | O | - | Not done |
| Isoform 1 of Mitotic checkpoint protein BUB3 (BUB3) | IPI00013468 (+2) | 37 | 2 | N.D. | 1 | O | O | - | Not done |
| ATP-dependent RNA helicase DDX1 (DDX1) | IPI00293655 (+1) | 82 | 2 | 1 | 1 | O | - | Δ [13] | Not done |
| Isoform 1 of Pogo transposable element with ZNF domain (POGZ) | IPI00410717 (+5) | 155 | 2 | 2 | N.D. | O | - | O [14] | Not done |
| Isoform PLZFA of Zinc finger and BTB domain-containing protein 16 (PLZF) | IPI00220823 (+1) | 62 | N.D. | 2 | N.D. | O | - | Δ [15] | Not done |
| Isoform Short of RNA-binding protein FUS (FUS) | IPI00221354 (+3) | 53 | 41 | 35 | 23 | O | - | Δ [16] | Not done |
| RNA-binding protein EWS isoform 1 (EWS) | IPI00009841 (+4) | 69 | 29 | 24 | 14 | O | - | Δ [17] | Not done |
| Proliferating cell nuclear antigen (PCNA) | IPI00021700 | 29 | N.D. | 1 | 3 | - | O | O [1] | O |
HP1γ, heterochromatin protein 1γ; M. W., molecular weight; N.D., not detected; O, validated by fold ratio, GO, or coimmunoprecipitation experiments (Co-IP Ex). In Article references, references indicated as O report its interaction with HP1γ and functional relation to DNA damage response; references indicated as Δ report only functional relation to DNA damage response.
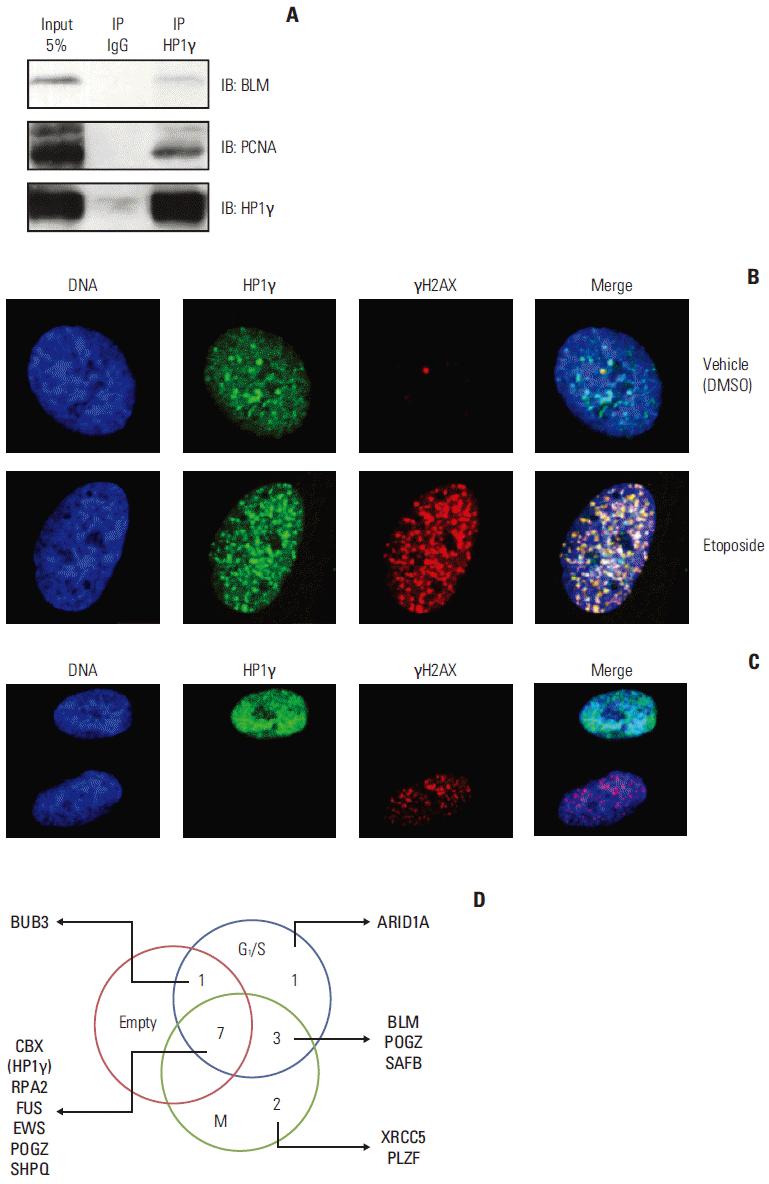
Fig. 3.




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download