Abstract
Anaplastic lymphoma kinase (ALK) fusion is a common mechanism underlying pathogenesis of non-small cell lung carcinoma (NSCLC) where these rearrangements represent important diagnostic and therapeutic targets. In this study, we found a new ALK fusion gene, SEC31A-ALK, in lung carcinoma from a 53-year-old Korean man. The conjoined region in the fusion transcript was generated by the fusion of SEC31A exon 21 and ALK exon 20 by genomic rearrangement, which contributed to generation of an intact, in-frame open reading frame. SEC31A-ALK encodes a predicted fusion protein of 1,438 amino acids comprising the WD40 domain of SEC31A at the N-terminus and ALK kinase domain at the C-terminus. Fluorescence in situ hybridization studies suggested that SEC31A-ALK was generated by an unbalanced genomic rearrangement associated with loss of the 3′-end of SEC31A. This is the first report of SEC31A-ALK fusion transcript in clinical NSCLC, which could be a novel diagnostic and therapeutic target for patients with NSCLC.
Point mutation and gene amplification events may enhance oncogenic activity of anaplastic lymphoma kinase (ALK); however, the most common genetic alteration involving ALK in tumors is genomic rearrangement [1]. Thus far, various translocations and inversions involving the 2p23 chromosomal locus harboring the ALK gene have been reported. These rearrangements result in fusion genes involving fusion of the 3ʹ-end of ALK to the 5ʹ-end of various partners [1]. The NPM-ALK chimeric gene, originally identified in patients with anaplastic large cell lymphoma, was the first reported fusion involving the ALK tyrosine kinase domain. Various other ALK chimeric proteins have recently been identified in several tumor types.
Interestingly, most oncogenic ALK fusion genes share common features. First, expression of the ALK fusion gene is regulated by the promoter of the 5ʹ-end partner gene, which usually encodes a protein widely expressed in healthy tissues [1]. Second, most N-terminal fusion partners contain domains that could be involved in oligomer formation, which is essential for the oncogenic activation of the ALK fusion protein [1]. Such oligomerization domains likely allow ligand-independent dimerization of ALK kinase domain and lead to constitutive kinase activation and aberrant activation of downstream signaling pathways for carcinogenesis [2,3].
Recent studies have demonstrated the important role of ALK in the pathogenesis of non-small cell lung cancer (NSCLC), where various ALK fusion genes have been identified. In particular, EML4-ALK [4] appeared as an oncogenic target in 6%-7% of NSCLC patients. In addition, kinesin family member 5B (KIF5B)–ALK, TRK-fused gene (TFG)– ALK, kinesin light chain 1 (KLC1)–ALK, protein tyrosine phosphatase and non-receptor type 3 (PTPN3)–ALK, huntingtin interacting protein 1 (HIP1)–ALK [5], and translocated promoter region (TPR)–ALK [6] have been found in lung adenocarcinoma by several research groups worldwide, including our group.
The oncogenic role of SEC31A-ALK fusion protein was well established in rare cases of ALK-positive large B-cell lymphomas [7,8] and inflammatory myofibroblastic tumor [9]; however, its occurrence in NSCLC has not been previously reported. Here, we report on the first case of SEC31A-ALK as an oncogenic target in NSCLC.
A 53-year-old Korean man was diagnosed with NSCLC. A computed tomography (CT) scan of the chest was performed, and an unusual mass measuring 1.5×1.0 cm2 was identified in the lower lobe of the right lung (Fig. 1A). Microscopic evaluation of the lesion showed a poorly differentiated adenocarcinoma (Fig. 1B and D). Immunohistochemistry for ALK (1:50, clone 5A4, Novocastra, Newcastle upon Tyne, UK) showed diffuse cytoplasmic and granular staining (Fig. 1E and F). Fluorescence in situ hybridization (FISH) analysis for ALK using the Vysis ALK FISH Breakapart Probe Kit (Abbott Molecular, Abbott Park, IL) showed clear splitting between the 5ʹ- and 3ʹ-probe signals (Fig. 1C). Fifteen months after lobectomy of the tumor mass, CT scans did not show any evidence of recurrence or metastatic disease.
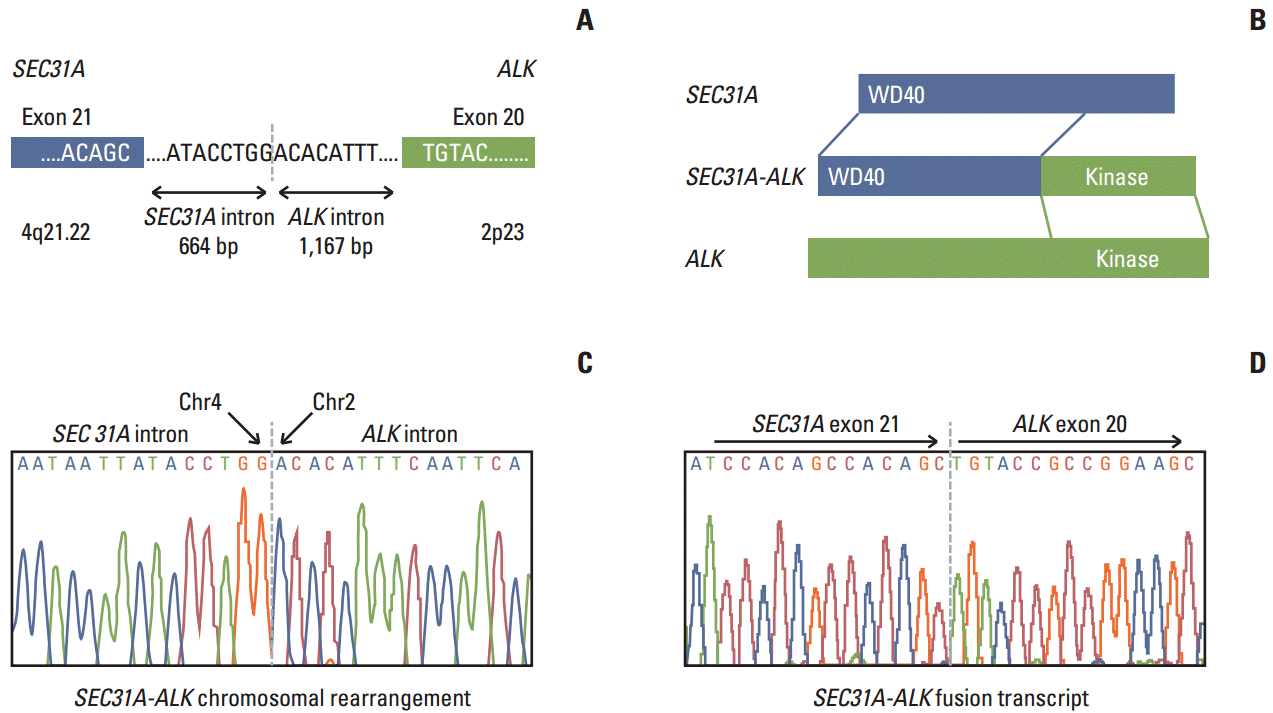
During screening for ALK fusion genes in the lung adenocarcinoma tumor using a single-tube multiplexed assay previously developed by our group [10], we found that the tumor tissue showed elevated expression of an ALK fusion transcript. Using Anchored Multiplex polymerase chain reaction (PCR) for ALK, RET, and ROS1 (Enzymatics Inc., Boulder, CO), followed by next generation sequencing on MiSeq (Illumina, San Diego, CA) and reverse transcription polymerase chain reaction, we found that the tumor expressed a novel SEC31A-ALK rearrangement bearing a fusion junction between SEC31A exon 21 and ALK exon 20, which could contribute to production of an intact, in-frame open reading frame (Fig. 2). Further genomic PCR showed that SEC31A-ALK occurred via genomic recombination between nucleotide 83764951 (nucleotide 664 downstream of the SEC31A exon 21) on chromosome 4 and nucleotide 29447585 (nucleotide 1167 upstream of the ALK exon 20) on chromosome 2 (Fig. 2).
FISH with SEC31A break-apart probes [7] showed one co-localized red and green signal and one red signal alone in neoplastic cells (Supplementary Fig. S1). This pattern indicated loss of the 3′-end of SEC31A covered by BAC clones labeled in SpectrumGreen (RP11-251L03/RP11-450H10/RP11-791G16/RP11-331E09) (Supplementary Fig. S2). The SEC31A-ALK fusion gene was eventually demonstrated using a combination of the SpectrumOrange-labeled 5′-end probes of SEC31A (RP11-163O17/RP11-93E17/RP11-66E15/RP11-353M17) and the SpectrumGreen-labeled 3′-end of ALK (P1 clone 1111H1) [11], which were co-localized in neoplastic cells (Supplementary Fig. S3). These FISH findings suggest that the SEC31A-ALK rearrangement was generated not by a reciprocal t(2;4)(p23;q21), but by complex and/or imbalanced genomic rearrangement, like those in the previously reported cases [7-9].
The new SEC31A-ALK fusion transcript described in this study encodes a predicted protein of 1,438 amino acids with a WD40 domain encoded by the 5ʹ-end of SEC31A and a tyrosine kinase domain encoded by the 3ʹ-end of ALK (Fig. 2A). Although this type of fusion gene was previously identified in large B-cell lymphoma and inflammatory myofibroblastic tumor (IMT), to the best of our knowledge, for the first time we discovered the SEC31A-ALK fusion gene in NSCLC. In contrast to the intact in-frame fusion between SEC31A exon 24 and ALK exon 20 in the previous fusion transcript identified in ALK-positive large B-cell lymphoma [7,8], the SEC31A-ALK fusion gene found in the reported case was formed by intact in-frame fusion between SEC31A exon 21 and ALK exon 20 at the fusion transcript level. However, all of these fusions harbored the same two intact functional domains (WD40 and tyrosine kinase), strongly suggesting that they play similar oncogenic roles [7,8]. We deposited the cDNA sequence for the new fusion transcript spanning the conjoined region at the transcript level into the NCBI Genbank under accession number KJ495955.
Lung cancer is the leading cause of cancer-related deaths in the world. ALK has recently emerged as a novel molecular target for therapeutic strategies in lung adenocarcinoma. Tumors with translocations involving ALK usually express chimeric transcripts containing the 3′-end region of ALK encoding tyrosine kinase domain and the 5′-end region of various partner genes [12]. The known ALK partner genes in lung adenocarcinoma include EML4 [4], TFG, KIF5B, KLC1, and PTPN3 [13]. Our research group recently reported two novel fusion genes, HIP1-ALK and TPR-ALK, in lung adenocarcinoma [5,6].
We report here on a new SEC31-ALK fusion variant identified in a case of ALK-positive lung adenocarcinoma. Our FISH assay with break-apart probes for ALK and SEC31A suggested that the chimeric gene was not generated by a reciprocal t(2;4)(p23.2;q21.22), but by an unbalanced rearrangement associated with loss of 3′ SEC31A.
The SEC31A gene, which is known to be ubiquitously expressed in human cells, encodes for a SEC31A (alias SEC31L1) protein found within vesicular structures, which are distributed throughout the cell and are especially c oncentrated at the perinuclear region. SEC31A is a component of the coat protein complexes involved in mediating transport from the endoplasmic reticulum to the Golgi complex [14]. Such cellular localization of SEC31A may explain a diffuse cytoplasmic and granular expression pattern of SEC31A-ALK observed in the current case of lung adenocarcinoma.
Oncogenic potential of SEC31A-ALK was previously demonstrated in vitro by Van Roosbroeck et al. [7], who showed that the fusion transforms interleukin 3 (IL-3)– dependent Ba/F3 cells to growth factor-independence and that kinase activity of SEC31A-ALK and its downstream signaling activity are reduced by NVP-TAE-684, the selective ALK inhibitor. In addition, suppression of Ba/F3 cell growth by NVP-TAE-684 was inhibited by treatment with IL3, thus reverting cell growth back to an IL-3 dependent pathway. The oncogenic tyrosine kinase activity of the SEC31A-ALK fusion protein is probably induced by the WD40 domain located at the N-terminus, which interacts with SEC13 and other proteins [9,14,15]. In the case of the SEC31L1-ALK variant identified in IMT, its fusion protein structure contained the same functional domains (WD40 and tyrosine kinase) [9].
SEC31A-ALK is a new ALK fusion gene in NSCLC. As previously demonstrated, the fusion harboring two intact functional domains, WD40 and ALK tyrosine kinase, is oncogenic and sensitive to NVP-TAE-684, an inhibitor of the ALK kinase [7]. Therefore, the SEC31A-ALK fusion gene is a potential driving rearrangement and diagnostic and therapeutic target for patients with NSCLC. In addition, our study confirms that NSCLC and ALK-positive lymphomas and IMT share the same ALK rearrangements.
Electronic Supplementary Material
Supplementary materials are available at Cancer Research and Treatment website (http://www.e-crt.org).
ACKNOWLEDGMENTS
This work was supported by grants from the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (MSIP) (NRF-2013M3C8A 1078501, NRF-2013R1A2A2A01068922 and No. 2011-0030001 for the Global Core Research Center [GCRC]).
References
1. Ardini E, Magnaghi P, Orsini P, Galvani A, Menichincheri M. Anaplastic lymphoma kinase: role in specific tumours, and development of small molecule inhibitors for cancer therapy. Cancer Lett. 2010; 299:81–94.

2. Webb TR, Slavish J, George RE, Look AT, Xue L, Jiang Q, et al. Anaplastic lymphoma kinase: role in cancer pathogenesis and small-molecule inhibitor development for therapy. Expert Rev Anticancer Ther. 2009; 9:331–56.

3. Drexler HG, Gignac SM, von Wasielewski R, Werner M, Dirks WG. Pathobiology of NPM-ALK and variant fusion genes in anaplastic large cell lymphoma and other lymphomas. Leukemia. 2000; 14:1533–59.

4. Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007; 448:561–6.

5. Hong M, Kim RN, Song JY, Choi SJ, Oh E, Lira ME, et al. HIP1-ALK, a novel fusion protein identified in lung adenocarcinoma. J Thorac Oncol. 2014; 9:419–22.

6. Choi YL, Lira ME, Hong M, Kim RN, Choi SJ, Song JY, et al. A novel fusion of TPR and ALK in lung adenocarcinoma. J Thorac Oncol. 2014; 9:563–6.

7. Van Roosbroeck K, Cools J, Dierickx D, Thomas J, Vandenberghe P, Stul M, et al. ALK-positive large B-cell lymphomas with cryptic SEC31A-ALK and NPM1-ALK fusions. Haematologica. 2010; 95:509–13.

8. Bedwell C, Rowe D, Moulton D, Jones G, Bown N, Bacon CM. Cytogenetically complex SEC31A-ALK fusions are recurrent in ALK-positive large B-cell lymphomas. Haematologica. 2011; 96:343–6.

9. Panagopoulos I, Nilsson T, Domanski HA, Isaksson M, Lindblom P, Mertens F, et al. Fusion of the SEC31L1 and ALK genes in an inflammatory myofibroblastic tumor. Int J Cancer. 2006; 118:1181–6.

10. Lira ME, Kim TM, Huang D, Deng S, Koh Y, Jang B, et al. Multiplexed gene expression and fusion transcript analysis to detect ALK fusions in lung cancer. J Mol Diagn. 2013; 15:51–61.

11. Mathew P, Sanger WG, Weisenburger DD, Valentine V, Pickering D, et al. Detection of the t(2;5)(p23;q35) and NPM-ALK fusion in non-Hodgkin's lymphoma by two-color fluorescence in situ hybridization. Blood. 1997; 89:1678–85.
12. Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010; 363:1693–703.
13. Ou SH, Bartlett CH, Mino-Kenudson M, Cui J, Iafrate AJ. Crizotinib for the treatment of ALK-rearranged non-small cell lung cancer: a success story to usher in the second decade of molecular targeted therapy in oncology. Oncologist. 2012; 17:1351–75.

14. Tang BL, Zhang T, Low DY, Wong ET, Horstmann H, Hong W. Mammalian homologues of yeast sec31p: an ubiquitously expressed form is localized to endoplasmic reticulum (ER) exit sites and is essential for ER-Golgi transport. J Biol Chem. 2000; 275:13597–604.
Fig. 1.
(A) Computed tomography scan of the chest showing a solid nodule with a speculated margin (arrow) measuring 1.5-cm in size in the lower lobe of the right lung. (B) Anaplastic lymphoma kinase (ALK) immunohistochemical staining showing cytoplasmic staining of tumor cells (×100). (C) Fluorescence in situ hybridization assay of ALK showing ALK genomic rearrangement by split 5'- and 3'-probe signals (arrows). (D-F) Hematoxylin and eosin staining showing adenocarcinoma (D, ×100), and a mixture of acinar pattern (E, ×200) and solid pattern (F, ×200).
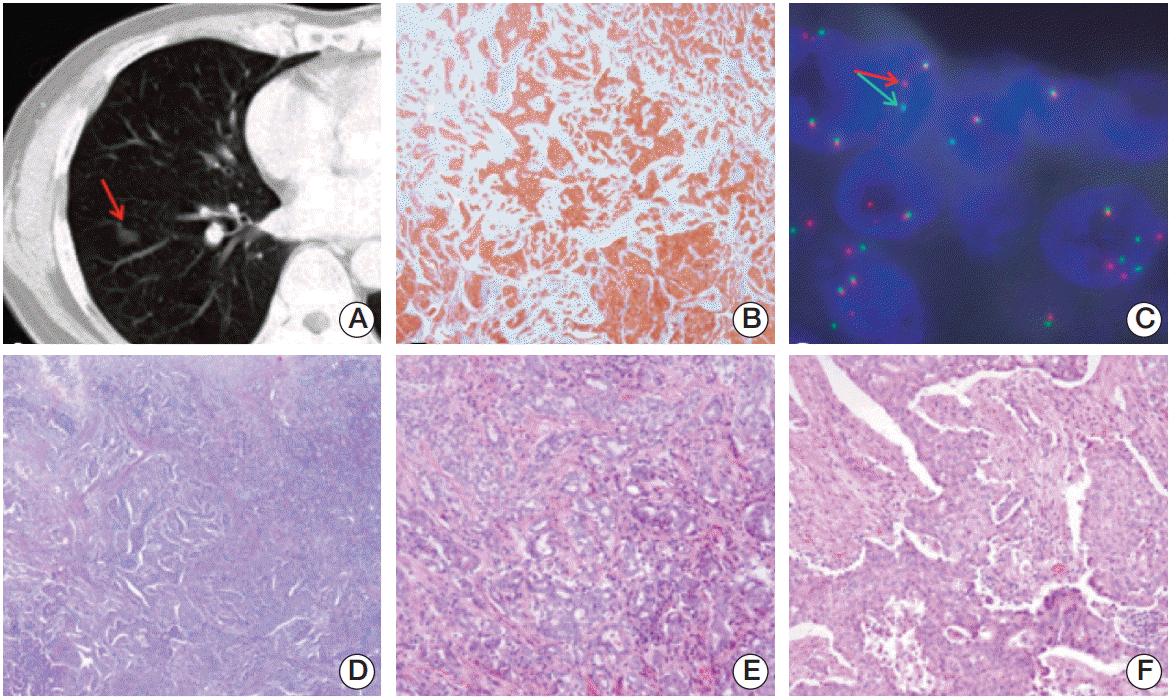
Fig. 2.
Schematic (A) showing the intron containing and surrounding the conjoined region for genomic rearrangement for the SEC31A-ALK fusion gene encoding the fusion protein (B) harboring the WD and tyrosine kinase functional domains as well as the adjacent exons. (C, D) DNA sequence chromatograms showing the conjoined regions at the genomic DNA sequence level (C) and cDNA sequence level (D) of the SEC31A-ALK fusion gene.




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download